Abstract
Rationale: Although the transplantation of induced pluripotent stem cell (iPSC)–derived cells harbors enormous potential for the treatment of pulmonary diseases, in vivo data demonstrating clear therapeutic benefits of human iPSC–derived cells in lung disease models are missing.
Objectives: We have tested the therapeutic potential of iPSC-derived macrophages in a humanized disease model of hereditary pulmonary alveolar proteinosis (PAP). Hereditary PAP is caused by a genetic defect of the GM-CSF (granulocyte–macrophage colony–stimulating factor) receptor, which leads to disturbed macrophage differentiation and protein/surfactant degradation in the lungs, subsequently resulting in severe respiratory insufficiency.
Methods: Macrophages derived from human iPSCs underwent intrapulmonary transplantation into humanized PAP mice, and engraftment, in vivo differentiation, and therapeutic efficacy of the transplanted cells were analyzed.
Measurements and Main Results: On intratracheal application, iPSC-derived macrophages engrafted in the lungs of humanized PAP mice. After 2 months, transplanted cells displayed the typical morphology, surface markers, functionality, and transcription profile of primary human alveolar macrophages. Alveolar proteinosis was significantly reduced as demonstrated by diminished protein content and surfactant protein D levels, decreased turbidity of the BAL fluid, and reduced surfactant deposition in the lungs of transplanted mice.
Conclusions: We here demonstrate for the first time that pulmonary transplantation of human iPSC–derived macrophages leads to pulmonary engraftment, their in situ differentiation to an alveolar macrophage phenotype, and a reduction of alveolar proteinosis in a humanized PAP model. To our knowledge, this finding presents the first proof-of-concept for the therapeutic potential of human iPSC-derived cells in a pulmonary disease and may have profound implications beyond the rare disease of PAP.
Keywords: induced pluripotent stem cell, cell therapy, pulmonary alveolar proteinosis, macrophages
At a Glance Commentary
Scientific Knowledge on the Subject
Induced pluripotent stem cells (iPSCs) have great therapeutic potential; however, data on actual in vivo therapeutic benefits of human iPSC–derived cells are scarce and are lacking completely with respect to pulmonary diseases. Hereditary pulmonary alveolar proteinosis (herPAP) is a rare, life-threatening lung disease caused by defective alveolar macrophages and characterized by massive lung proteinosis. No causal therapy is available, but marked and long-lasting therapeutic benefit of intrapulmonary transplanted macrophages has been demonstrated. Importantly, in contrast to other pulmonary cell populations, the effective generation of macrophages from human iPSC sources already has been established.
What This Study Adds to the Field
Employing a humanized herPAP model, we here demonstrate that on intrapulmonary transplantation human iPSC–derived macrophages develop into alveolar macrophage–like cells, significantly reducing alveolar protein and surfactant deposition and profoundly ameliorating the herPAP phenotype. To the best of our knowledge, this study represents the first proof-of-concept for the therapeutic efficacy of human iPSC-derived cells in a pulmonary disease and illustrates the potential of iPSCs and progeny derived thereof in respiratory diseases. These data should have considerable implications for the future treatment of herPAP as well as other hereditary and acquired lung diseases.
Induced pluripotent stem cells (iPSCs) are reprogrammed somatic cells genetically set back to a pluripotent state, thereby allowing for theoretically unlimited expansion and differentiation into any cell type of the human body (1, 2). iPSCs hold great promise to generate cells for patient-specific, nonimmunogenic gene and cell therapy approaches. So far, numerous iPSC-based cell types and in vitro disease models have been developed (3–7); however, data on the actual in vivo therapeutic benefit of human iPSC–derived cells remain scarce (8).With respect to the lungs, the generation of functional pulmonary cell populations from pluripotent cells has been cumbersome, although some progress on the successful generation of human lung progenitor and human lung epithelial cells (9–11) from iPSC sources has been reported. More success has been reported in the generation of functional human macrophages including alveolar macrophage (AM)-like cells (12–14). However, demonstration of a therapeutic benefit of human iPSC–derived cells in a lung disease model is still pending. In our current work, we therefore evaluated the therapeutic potential of human iPSC–derived macrophages in a humanized mouse model of hereditary pulmonary alveolar proteinosis (herPAP).
herPAP is a congenital disease caused by recessive mutations in the GM-CSF (granulocyte–macrophage colony–stimulating factor) receptor A or B gene (CSF2RA and CSF2RB), leading to insufficient AM differentiation and function (15–17). The inability of pulmonary alveolar proteinosis (PAP)-AMs to degrade pulmonary proteins and phospholipids results in massive lung proteinosis, followed by disturbed gas exchange and life-threatening respiratory insufficiency. The only therapy currently available is repetitive whole-lung lavage under full anesthesia, a purely symptomatic, burdensome, and dangerous procedure for the patient (18). New causal and long-lasting therapies are urgently needed to improve the quality of life, and life expectancy, of affected patients. In this context we and partners have reported on the therapeutic benefit of pulmonary macrophage transplantation (PMT) in herPAP (19, 20).
In this background, the aim of the current study was to test whether iPSCs can serve as an effective source for the generation of AMs that can functionally replace the defective macrophage population in patients with PAP and efficiently degrade the accumulated alveolar proteins and phospholipids. In this respect, we investigated the intrapulmonary transplantation of human iPSC–derived macrophages in an established humanized animal model of herPAP and describe effective homing and engraftment of iPSC-derived transplanted macrophages in the recipient lungs, followed by their differentiation into functional AM-like cells within the lung environment, resulting in degradation of the accumulated alveolar proteins. To the best of our knowledge, these data demonstrate for the first time the therapeutic potential of human iPSC–derived cells in an established in vivo lung disease model.
Methods
Culture and Hematopoietic Differentiation of Human iPSCs
The human iPSC clone (hCD34iPSC16) was cultured and differentiated as previously described (19). In brief, human iPSCs were cultured on irradiated murine CF1 feeder cells and knockout Dulbecco’s modified Eagle’s medium supplemented with 20% knockout serum replacement, 1 mM l-glutamine, 1% nonessential amino acids, 1% penicillin–streptomycin, 0.1 mM β-mercaptoethanol, and bFGF (basic fibroblast growth factor) (20–40 ng/ml) (12). Human iPSCs were subjected to embryoid body formation for 5 days in the presence of 10 μM ROCK (Rho-associated, coiled coil–containing protein kinase) inhibitor (Y-27632) and then manually transferred to 6-well adherent plates and cultured for 7 days in X-VIVO 15 medium supplemented with 1% penicillin–streptomycin, human IL-3 (hIL-3; 25 ng/ml), and human M-CSF (macrophage–colony stimulating factor) (hM-CSF; 50 ng/ml). Starting from Day 10–15, cells from differentiation cultures were harvested once weekly from the supernatant. For transplantation experiments, cells were used directly after harvesting.
Colony Formation Assay
Human iPSC–derived macrophages (1–50 × 103) were cultured in triplicate in methylcellulose-based colony-forming assays supplemented with human cytokines (human kit #H4434; STEMCELL Technologies). After 10–12 days, colonies containing more than 50 cells were scored, using an inverted microscope.
Mice and Treatment Protocol
Humanized pulmonary alveolar proteinosis (huPAP) [129S4-Rag2tm1.1Flv Csf2/Il3tm1.1(CSF2,IL3)Flv Il2rgtm1.1Flv/J] mice (21) were obtained from Jackson Laboratory and housed in the local central animal facility of Hannover Medical School. NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJZtm) were obtained from the local central animal facility. Mice were kept in individually ventilated cages and had free access to food and water. Treated mice received intrapulmonary transplantations of 1–4 × 106 human iPSC–derived macrophages per week. For this, they received brief anesthesia and were orally intubated. Cells were applied intratracheally in a volume of 50–75 μl of phosphate-buffered saline (PBS). Two months after the first treatment, mice were sectioned and their phenotype was compared with that of untreated control animals. Experiments were performed two or three times with age- and sex-matched mice within treated and untreated huPAP groups.
Collection of Murine BAL Fluid and Measurement of Protein Amount and Surfactant Levels
Murine BAL was performed by cannulating the murine trachea postmortem, and lavaging the right lung three times with 400 μl of PBS. BAL fluid (BALF) was stained for flow cytometry or immediately frozen to −80°C and stored until analysis of protein concentration (Pierce BCA protein assay kit; Thermo Scientific). BALF surfactant levels were determined as previously described (19).
Preparation of Lung Homogenates
Lung homogenates were prepared by blending into less than 1-mm pieces and incubation with collagenase–DNase for 45 minutes at 37°C (gentleMACS kit; Miltenyi Biotec) followed by repeated mincing, filtering through a 70-μm cell strainer (Becton Dickinson), red blood cell lysis, and extensive washing.
Cell Isolation and Collection of Human Material
iPSC- and CD34-derived cells from transplanted organs were sorted by means of flow cytometry in the central sorting facility at Hannover Medical School. To discriminate cell-specific signals from autofluorescent lipid droplets in the lungs of huPAP mice, human cells were stained for hCD45 in allophycocyanin and sorted with a MoFlo flow cytometer (Beckman Coulter). Human BAL collection and cell isolation were conducted according to local standard operation procedures. To obtain pure primary AMs, Percoll isolation (22) of BALF cells from pediatric patients with lung inflammation was done; purity was checked by cytospins and was usually above 98%. For in situ analyses of primary human AMs, an unused piece of healthy human donor lung allocated from transplantation medicine was used. The study was approved by local institutional review boards, and all study subjects or their legal guardians gave written informed consent, and minors gave assent.
RT-PCR
Total RNA was isolated by use of an RNeasy mini or micro kit (Qiagen), according to the manufacturer’s instructions. Reverse transcription was performed with oligo(dT) primers and a complementary DNA reverse transcription kit (Fermentas). Complementary DNA (50 ng) was used for RT-PCR with PCRMasterMixGold (Peqlab) and primers for human GAPDH or mouse β-actin (Hs_GAPDH_2_SG and b-actin Mm_Actb_2_SG, respectively; both from Qiagen). Results were analyzed by agarose gel electrophoresis.
Karyotyping
Chromosome preparation and fluorescence R-banding were performed as previously described (23). Karyotypes are annotated according to the International System for Human Cytogenetic Nomenclature (2009).
Chip Cytometry
Multiplex chip cytometry was performed as described previously (19, 24) (http://www.zellkraftwerk.com). Primary murine and donor-derived cells were fixed on chips and processed with a ZellScanner ONE (Zellkraftwerk). For tissue sections, fixed and cryopreserved lungs were cut with a CM1950 cryostat (Leica), and 3-μm thin sections were fixed on a chip. Antibodies (Table E2 in the online supplement) were incubated for 10–15 minutes at room temperature. Cell-individual fluorescence signals were subtracted from respective background pictures. Principal component analyses were created with the R programming language.
Murine BALF Surfactant Clearance Assay
iPSC-derived macrophages were prestimulated for 1 week with GM-CSF (50 ng/ml). Thereafter, they were incubated with murine huPAP BALF for 24 hours and then either fixed and stained or washed to remove residual surfactant and cultured for an additional 24 hours. Fixation was performed with 10% formalin solution followed by oil red O (Sigma-Aldrich) staining. Briefly, fixed cells were incubated for 2 minutes in isopropanol and stained with freshly prepared oil red O staining solution for 15 minutes followed by rinsing with water. Nuclei were stained by hematoxylin for 2 minutes and washed in warm water.
Histologies and Cytospins
For histological analyses, lungs were fixed in 4% methanol-free paraformaldehyde overnight, followed by a 12-hour incubation in 30% sucrose–PBS. Lung tissue was then frozen to −80°C in mounting medium and subsequently cut with a CM1950 cryostat (Leica). Thin sections (3 μm) were fixed on slides and stained with periodic acid–Schiff reagent (PAS). Histological stainings were photographed with an AxioCam ICc 1 camera and further analyzed with AxioVision software version 4.8.2 (Zeiss) and in-house programs. Cytospins were performed as preciously described (12, 19).
RNA Sequencing and Data Analysis
For a detailed description of transcriptomics methods, refer to the online supplement. To generate RNA libraries, 1 ng of total RNA per sample was used for library preparation with the SMARTer stranded total RNA-Seq kit—pico input mammalian (635006; Takara/Clontech). Libraries were barcoded by dual indexing and fragment length distribution was monitored with a Bioanalyzer high-sensitivity DNA assay (Agilent Technologies). Quantification of libraries was performed by use of the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). For NextSeq 500 sequencing, equal molar amounts of individual libraries were pooled, denatured with NaOH, and diluted to 1.5 pM (Illumina). A 1.3-ml volume of denatured pool (containing 1% of PhiX control) was run on an Illumina NextSeq 500 sequencer using a high-output flowcell for 75-bp single reads (#FC-404-2005; Illumina). For raw data processing and quality control, binary base call (BCL) files were converted to FASTQ files, using bcl2fastq conversion software version 2.16.0.10 (Illumina), and further trimmed using Trim Galore! (version 0.4.1). FASTQ files were mapped against a reference genome with the splice-aware aligner Spliced Transcripts Alignment to a Reference (STAR) (version 2.5.0c), and the resulting binary alignment map (BAM) files were built in a two-pass mapping process and sorted. Genome index files were created by STAR with default settings, using Homo sapiens sequence and annotation data (Ensembl, build GRCh37). For quantification, final data analysis, and visualization, BAM files were imported to StrandNGS (version 2.6). Quantification was performed by DESeq normalization, and baseline transformation to the mean of all analyzed samples was finally conducted.
Statistics
Graphs were created with Prism V5 (GraphPad Software), and statistical analysis was performed with Prism V6 (GraphPad). For calculation of significant differences in experiments with two groups, unpaired Student’s t testing was applied. For experiments with more than two groups, depending on data structure, one-way ANOVA with Tukey post hoc testing or Kruskal-Wallis with Dunn’s post hoc testing was applied. Significance is indicated with asterisks: *P < 0.05; **P < 0.01; ***P < 0.001.
Animal Study Protocol
All animal experiments were approved by the Lower Saxony State animal welfare committee (approval No. 12/0959) and performed according to their guidelines.
Results
We hypothesized that iPSC-derived human macrophage progenitors can be an effective source for intrapulmonary cell therapy in PAP and tested our hypothesis in huPAP mice [129S4-Rag2tm1.1Flv Csf2/Il3tm1.1(CSF2,IL3)Flv Il2rgtm1.1Flv/J]. huPAP mice are generated on the immunodeficient Rag2−/− Il2rg−/− background and carry a targeted replacement of the regions encoding murine IL-3 and GM-CSF (mIL-3 and mGM-CSF) with those encoding human IL-3 and human GM-CSF (hIL-3 and hGM-CSF). Because hGM-CSF and hIL-3 are not cross-reactive with mGM-CSF and mIL-3 (25), loss of mGM-CSF leads to a block of inherent AM differentiation and a fully developed herPAP phenotype, including high serum and BALF levels of hGM-CSF. These high hGM-CSF levels facilitate engraftment and differentiation of transplanted human cells in huPAP mice (21).
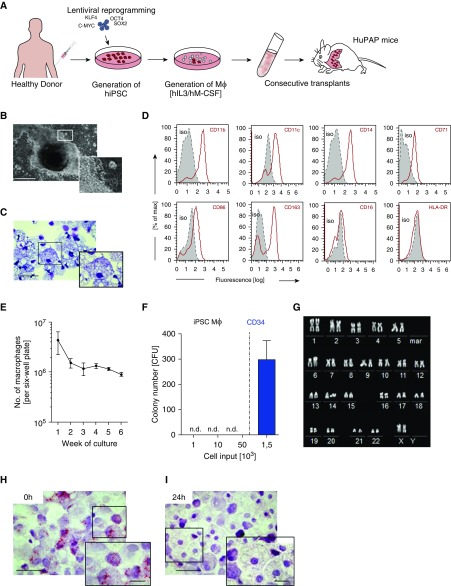
To generate large numbers of human iPSC–derived macrophages for in vivo experiments, we employed our recently developed three-step embryoid body–based differentiation protocol (Figures 1A and 1B). This robust and highly reproducible hematopoietic differentiation protocol allows the efficient generation of mature and functional macrophages from human iPSC sources (12). The macrophages obtained recapitulated the typical macrophage morphology (Figure 1C and Figure E1a in the online supplement) and phenotype with a CD45+CD11b+CD163+CD71+CD14+CD16−CD86+HLA-DR− surface marker pattern (Figure 1D), whereas surface markers characterizing human pluripotent stem cells (TRA1–60 [tumor related protein]), multipotent stem cells (CD34), B cells (CD19), or granulocytes (CD66b) were absent (12, 26).
Figure 1.
Generation and characterization of induced pluripotent stem cell (iPSC)-derived macrophages. (A) Generation of human iPSCs: Hematopoietic differentiation of iPSC-derived macrophages (iPSC Mϕ) with human macrophage colony–stimulating factor and human IL-3 and transplantation into humanized pulmonary alveolar proteinosis (HuPAP) mice. (B) Continuous generation of human iPSC Mϕ from embryoid body–derived structures (bright-field image; scale bar, 500 μm). (C) Morphology of iPSC Mϕ after cytospin preparation and May-Giemsa staining (scale bar, 50 μm). (D) Flow cytometric analysis of surface marker expression in iPSC Mϕ (gray, respective isotype control; red, stained surface marker; n = 3 experiments). (E) Overall numbers of macrophages from human iPSCs, generated in a six-well plate over 6 weeks of differentiation (n = 6, mean ± SEM). (F) Methylcellulose-based clonogenic assays performed with either iPSC Mϕ with various cell numbers or CD34+ cells derived from umbilical cord blood (n = 3, mean + SEM). (G) Chromosomal analysis of iPSC-derived macrophages by spectral R-banding. (H and I) Oil red O staining of iPSC-derived macrophages after incubation with BAL fluid from HuPAP mice (0 h). Cells were then washed and further cultured to demonstrate clearance of surfactant material after 24 hours (scale bars, 50 μm). C-MYC = MYC proto-oncogene; CFU = colony-forming unit; hIL3 = human IL-3; hiPSC = human induced pluripotent stem cell; hM-CSF = human macrophage colony–stimulating factor; KLF4 = Kruppel-like factor 4; mar = marker; n.d. = not detected; OCT4 = octamer-binding transcription 4; SOX2 = sex-determining region Y-box 2.
The differentiation protocol yielded stable cell numbers over 6 weeks, with 2–3 × 106 macrophages per plate per week (Figure 1E). The iPSC-derived macrophages completely lacked clonogenic growth potential (Figure 1F and Figure E1b) and did not display chromosomal abnormalities (Figure 1G) or aberrant γH2AX+ foci that could indicate DNA double-strand breaks (27) (Figure E1c). In vitro, these iPSC-derived macrophages demonstrated cell type–specific functionality and effectively phagocytosed and degraded murine BALF–derived lipoproteins (Figures 1H and 1I). Cells generated by this protocol also display other macrophage typical functionalities such as the phagocytosis of gold and latex particles or the clearance of GM-CSF from cell supernatants (19).
To test the in vivo therapeutic potential of these cells, human iPSC–derived macrophages were transplanted intratracheally into huPAP recipient mice (once weekly over 4 wk). Engraftment, cell localization, cell differentiation, and therapeutic efficiency were analyzed 2 months after the first transplantation.
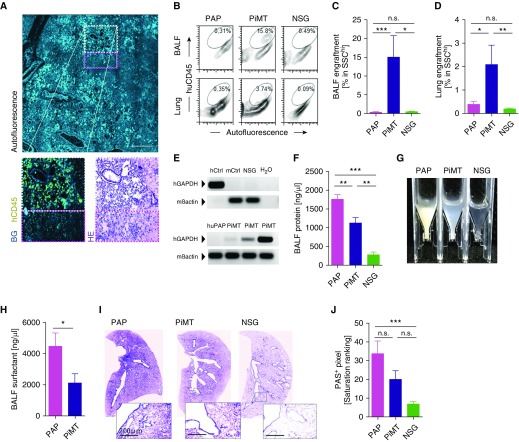
In treated animals, huCD45+ cells were detected in the lungs of recipient animals by immunofluorescence lung tissue stainings and were located near the large airways (Figure 2A). Flow cytometry further confirmed this engraftment and revealed a clear population of highly granular human (SSChi huCD45+) BALF and lung cells after transplantation (Figures 2B–2D and Figure E2a), whereas extrapulmonary engraftment was not detectable by flow cytometry in bone marrow, liver, or spleen (Figure E2b). RT-PCR further confirmed human cell engraftment in the lungs of transplanted mice (Figure 2E). In contrast to flow cytometry analyses, however, low amounts of human RNA were traceable in the spleens of treated mice by RT-PCR, potentially indicating splenic localization of iPSC-derived macrophages or progeny derived thereof, although at lower numbers (Figure E2c), demonstrating that the engraftment was not completely lung specific. Although the data presented here employed repetitive pulmonary iPSC-based macrophage transplantation (PiMT), we also observed successful engraftment and therapeutic benefit with single transplants in some mice (Table E1 and Figure E3).
Figure 2.
Engraftment and clinical benefit of human induced pluripotent stem cell (iPSC)–derived macrophages in humanized pulmonary alveolar proteinosis (huPAP) mice analyzed 2 months after the first transplantation. (A) Detection of human CD45+ cells within the lungs of recipient mice (light blue, autofluorescence; yellow, human CD45 [hCD45, huCD45]), with less consolidated alveoli in regions with cell engraftment (white dashed frame) compared with adjacent regions where no cells occurred (pink dashed frame) (scale bar, 250 μm). (B) Flow cytometry of BAL fluid (BALF) and lung cells reveals clear population of hCD45+ cells within the SSChi gate after pulmonary iPSC-based macrophage transplantation (PiMT) but not in untreated (PAP) or untransplanted control nonobese diabetic severe combined immunodeficient (NSG) mice. (C and D) BALF and lung chimerism as assessed by flow cytometry analyzing human CD45+ cells (n = 8 mice in PAP group, n = 10 in PiMT group, and n = 5 in NSG group; pooled data from three experiments). (E) Detection of human GAPDH in lungs of huPAP mice transplanted with iPSC-derived macrophages (PiMT, three mice). Primers detecting murine β-actin (mβactin) were used to detect murine DNA. Lungs of nontransplanted huPAP mice (huPAP) or NSG mice (NSG), a human cell line (hCtrl) or murine liver samples (mCtrl) were used as controls. (F) Reduced BALF protein levels (n = 10 mice in PAP group, n = 9 in PiMT group, and n = 9 in NSG group; pooled data from three experiments). (G) Reduced BALF turbidity after PiMT (photograph of representative BALF flushes). (H) Reduced BALF surfactant-D levels (n = 5 mice in PAP group, n = 6 in PiMT group; pooled data from two experiments). (I) Staining for periodic acid–Schiff reagent (PAS)-positive mucus in lung slices of treated and untreated mice (scale bars, 200 μm). (J) Reduced PAS-positive pixels after PiMT (n = 8 mice in PAP group, n = 10 in PiMT group, and n = 10 in NSG group; pooled data from three experiments). All graphs display mean + SEM; one-way ANOVA testing for three-group or one-tailed t test for two-group comparison: *P < 0.05, **P < 0.01, ***P < 0.001. BG = background; HE = hematoxylin and eosin; hGAPDH = human GAPDH; n.s. = not significant; SSC = side scatter.
To assess the efficacy of our new therapeutic approach in the humanized PAP disease model, we focused on readouts that are used in the clinical assessment of patients suffering from herPAP. Here, the gold standard for the diagnosis of herPAP is the demonstration of high BALF turbidity and protein content as well as increased intraalveolar deposition of proteins and phospholipids in specimens obtained from transbronchial lung biopsies (28). Also, the success of repetitive BAL, currently the only available therapy for herPAP, is routinely evaluated by measuring turbidity and protein content of the first and last portion of BALF from the patient. In our humanized murine model, the engraftment of huCD45+ cells was associated with a significant improvement of alveolar proteinosis in huPAP mice. Two months after transplantation, BALF protein content and BALF turbidity were significantly reduced (Figures 2F and 2G). Furthermore, the amount of surfactant protein D, the primary airway lipoprotein elevated in human and murine PAP, was significantly diminished after cell therapy, and PAS positivity of lung cryosections reflecting alveolar surfactant load was improved (Figures 2H–2J). All animals tolerated the treatment well, and we did not observe any side effects after transplantation. Specifically, there was no evidence of teratoma or tumor formation in the lungs or other organs of recipient mice on careful histological analysis.
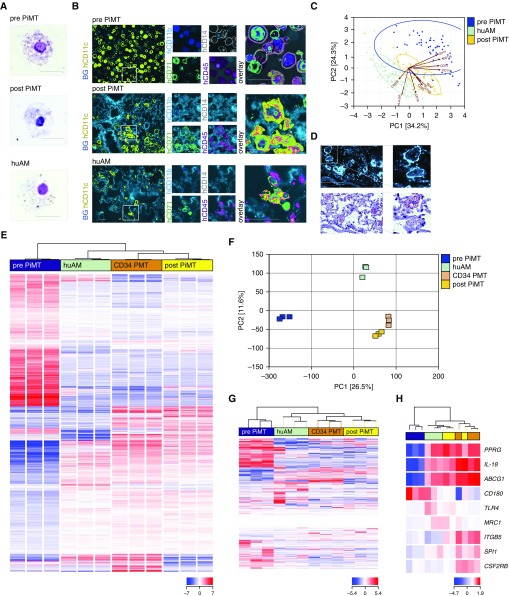
Next, we were interested in the in vivo differentiation and fate of human iPSC–derived macrophages after intrapulmonary transplantation into huPAP mice. In cytospins, huCD45+ cells sorted from the lungs of recipient animals resembled the appearance of primary human AMs (Figure 3A). Whereas freshly cultivated iPSC-derived macrophages were CD45+CD11c+CD71+CD11b+CD14+, macrophages isolated from the lungs of cell recipients 8 weeks after first cell transplantation remained CD45+CD11c+CD71+ but became CD11blowCD14−, demonstrating their adaptation to the marker panel of primary human AMs and thereby their adaptation to the lung microenvironment (Figure 3B). This finding was further corroborated in a principal component analysis of 10 surface markers that clearly demonstrated a change in the surface marker profile of iPSC-derived macrophages before versus after transplantation, when the iPSC-derived macrophages had become more similar to mature primary human AMs (Figure 3C). Moreover, histological stainings of lung cryosections of cell recipients revealed fine oil red O speckling of human CD45+ cells, indicating successful surfactant uptake as an essential AM function in lung physiology (Figure 3D).
Figure 3.
Phenotypic changes and protein and gene expression of induced pluripotent stem cell (iPSC)–derived macrophages 2 months after pulmonary transplantation therapy. (A) Cytospin appearance of iPSC-derived macrophages before and after transplantation (pulmonary iPSC-based macrophage transplantation [PiMT]) compared with primary human alveolar macrophages (huAMs; scale bar, 50 μm; representative experiment of three). (B) Surface protein expression and (C) principal component analysis (PCA) of 10 surface markers analyzed by chip cytometry of cells before and after PiMT, compared with the phenotype of human primary AMs within a lung specimen of a healthy human organ donor (representative experiment of two). Scale bar in B, 20 μm. (D) In situ staining of recipient lung tissue for human CD45 and oil red O staining of respective cells reveal intracellular lipoprotein uptake within recipient humanized pulmonary alveolar proteinosis (huPAP) lungs (scale bar, 50 μm; representative experiment of two). (E) Hierarchical clustering of 2,381 transcripts differentially expressed in the analyzed cell populations. One-way ANOVA testing was conducted to identify transcripts showing the most reliable mRNA expression differences in any of the possible pairwise comparisons between cells before (blue; pre-PiMT) and after transplantation (yellow; post-PiMT), primary huAMs (green) and alveolar macrophage–like cells reisolated from huPAP lungs 5 months after CD34 cell–based pulmonary macrophage transplantation (orange; CD34 PMT) (corrected P value < 0.01 according to Benjamini-Hochberg correction). Samples and transcripts were hierarchically clustered (distance metric, Euclidean; linkage rule, Ward’s). Red color indicates elevated relative mRNA expression levels, whereas blue color represents diminished mRNA levels. (F) PCA was conducted with unfiltered quantified RNA-Seq data. A two-dimensional representation focusing on main components 1 (26.52% of variance) and 2 (11.55% of variance) is depicted. (G and H) Hierarchical clustering of selected gene sets for (G) iPSC-determining and (H) AM-relevant genes, further illustrating the changes of iPSC-derived macrophages into AM-like cells after PiMT (at least three biological replicates from at least two independent experiments per cell type). ABCG1 = ATP binding cassette subfamily G member 1; BG = background; CSF2RB = granulocyte–macrophage colony–stimulating factor 2 receptor B; hCD45 = human CD45; ITGB5 = integrin β-5; MRC1 = mannose receptor 1; PC = principal component; PPRG = peroxisome proliferator–activated receptor-γ; SPI = spleen focus forming virus (SFFV) proviral integration oncogene (transcription factor PU.1); TLR4 = Toll-like receptor 4.
To further investigate the influence of the lung environment on human iPSC–derived macrophages, we compared the transcription profiles of four different groups of cells: 1) iPSC-derived macrophages before transplantation (pre-PiMT), 2) iPSC-derived macrophages 2 months after transplantation (post-PiMT), 3) bone marrow–derived macrophages 5 months after transplantation (CD34 PMT), and 4) primary human AMs (huAMs) isolated from BAL fluid of humans (huAMs).
In unbiased hierarchical cluster analysis of gene expression data of these various macrophage types, human iPSC–derived macrophages after transplantation (post-PiMT), bone marrow–derived macrophages after transplantation (CD34 PMT), and primary human AMs (huAMs) displayed similar expression patterns that clearly differed from those of iPSC-derived macrophages before transplantation (pre-PiMT) (Figure 3E). This profound change in overall gene expression of iPSC-derived macrophages after transplantation in the microenvironment of the lung was also confirmed by principal component analysis of whole transcriptome data (Figure 3F), where human AMs (huAMs), iPSCs (post-PiMT), and BM-derived macrophages (CD34 PMT) after transplantation clustered together and were different from iPSC-derived macrophages before transplantation (pre-PiMT). Furthermore, hierarchical cluster analyses of a selected gene set associated with pluripotency (29) yielded a similar clustering pattern, with marked downregulation of pluripotency genes in the iPSC-derived macrophages after transplantation (post-PiMT) compared with iPSC-derived macrophages before transplantation (pre-PiMT) (Figure 3G). A comparable result was observed after enrichment for genes relevant for GM-CSF–induced macrophage maturation (19). Similar to primary human AMs and CD34+ cell–derived macrophages after transplantation (CD34 PMT), iPSC-derived cells after transplantation (post-PiMT) displayed a marked upregulation of PPRG (peroxisome proliferator–activated receptor-γ), a negative regulator of macrophage activation (30), and macrophage-derived factors limiting lung inflammation such as IL-18 and ABCG1 (ATP-binding cassette transporter G1) (31, 32). Interestingly, specific factors that are known to drive terminal GM-CSF–triggered macrophage differentiation/activation, such as the transcription factor PU.1 (SPI1) and the β chain of the GM-CSF receptor (CSF2RB), were more highly expressed in iPSC-derived macrophages compared with primary human AMs and most similar to the expression in cells reisolated after CD34 cell–based PMT to huPAP mice (Figure 3H).
Discussion
This study presents clear evidence for the in vivo therapeutic efficacy of intrapulmonary transplantation of human iPSC–derived macrophages in a well-established disease model of a hereditary lung disease. In our humanized herPAP model, human iPSC–derived cells were detected within the recipient lungs for at least 2 months after transplantation. Human cells recovered at this time point from the recipient BALF or lungs displayed profound changes in phenotype and gene expression profile when compared with the transplanted cell population. Moreover, critical disease parameters such as bronchoalveolar protein content, surfactant protein D levels, and turbidity as well as alveolar deposition of PAS-positive protein and surfactant material were clearly reduced in transplanted PAP mice.
These data are in line with our concept of PMT as an effective treatment approach to herPAP. We and partners previously showed that intratracheal application of healthy or genetically corrected bone marrow–derived macrophages allows for the substitution of deficient PAP macrophages by an AM-like population capable of efficient phagocytosis and elimination of accumulated lipoproteins (19, 20). In contrast to allogeneic bone marrow transplantation or hematopoietic stem cell–based gene therapy approaches, PMT can be performed without prior chemo- and/or radiotherapeutic conditioning of the recipient. This constitutes a crucial advantage of the procedure. Even though successful, hematopoietic stem cell–based gene therapy has been demonstrated in Csf2Rb-deficient mice (33, 34), in the clinical setting the severely damaged lungs of patients with PAP tolerate the conditioning procedure poorly (16).
Whereas in our previous studies primarily bone marrow–derived macrophages were employed for PMT, the use of iPSCs in this context would enable large-scale culture and potentially unlimited expansion of patient-specific, gene-corrected macrophages, thus representing an extremely promising tool for future clinical cell therapies (35, 36). Along this line, we and others have developed a robust and highly effective protocol for the large-scale generation of monocytes/macrophages from pluripotent cell sources (12, 14, 37). Using this technology, we are able to generate well above 5 × 108 cells, corresponding to the cell numbers necessary for human cell therapy as extrapolated from our murine studies (19, 20).
In our work, we analyze the novel therapeutic concept of intrapulmonary cell therapy for herPAP, employing human iPSC–derived macrophages in humanized PAP mice. These mice are generated on the immunodeficient Rag2−/− Il2rg−/− background and lack T, B, and natural killer cells, rendering them effective recipients for human cells (21). However, in this model important human homeostatic mechanisms such as cell niche interactions are not present, a general and well-recognized limitation of humanized mouse models. In this context, we refer to our previous work with murine cells in congenic immunocompetent mice, which more closely resemble the situation of autologous cell transplantation in the clinical setting (19, 20). This congenic transplantation model yielded effective engraftment and therapeutic benefit, further supporting the feasibility of intrapulmonary cell transplantation in patients with herPAP.
Similar to our approach, in a murine system, the generation of a functional, in vitro–expandable population of AM-like cells from pluripotent stem cells has been described. These cells were able to engraft in immunodeficient adenosine deaminase–deficient mice, as well as rescue these animals from respiratory failure and promote pulmonary epithelial repair on intrapulmonary transplantation (13).
Interestingly, within 2 months in the lung environment, the phenotype and genotype of our transplanted cells closely adapted to those of human AMs, as demonstrated by surface marker and transcriptome analysis. These data fit the current concept of AM biology, which suggests that tissue-resident macrophages (TRMs) differentiate from early precursors in the yolk sac blood islands or fetal liver in response to the local microenvironment (38–40). In several studies these precursors were linked to the primitive wave of hematopoiesis, which is independent of c-myb expression and distinct from the generation of reconstituting hematopoietic stem cells by definite hematopoiesis (39, 41, 42). Similar to other TRM populations, under physiological conditions AMs self-sustain throughout adult life, and only after loss of the primary TRM population due to disease or damage (e.g., radiation, infections) are significant amount of TRMs replenished via peripheral blood monocytes (38). Moreover, TRM populations are characterized by substantial plasticity. This has been demonstrated by the transfer of distinct primary TRM populations into new tissue and organ microenvironments (43). Such plasticity might also facilitate the application of iPSC-derived macrophages in other macrophage- or TRM-related diseases such as heme oxygenase-1 deficiency or adrenoleukodystrophy (44, 45).
Our studies suggest that autologous, gene-corrected, iPSC-derived macrophages represent a potential therapeutic source for patients with the hereditary form of pulmonary alveolar proteinosis. This approach requires the generation of patient-specific iPSCs and their genetic correction and expansion before intrapulmonary reapplication to the specific patient. To this point, gene correction of iPSCs derived from patients with PAP already has been demonstrated to reverse, at least in vitro, not only the PAP phenotype but also the associated functional defects (7, 19). Although this work employed a lentiviral vector–based gene addition approach, modern designer nucleases even allow for the site-specific correction of defective genes, thus preserving physiological gene regulation and avoiding the risk of insertional mutagenesis (46). Moreover, so-called “universal” donor iPSC lines genetically engineered for low or absent expression of HLA molecules have been described (47, 48). Although the lack of HLA expression certainly reduces immunorejection of these cells and facilitates allogeneic transplantation scenarios even in the absence of immunosuppression, the effect of HLA knockdown on the functional properties of macrophage populations remains to be determined.
We consider our work as a first proof-of-concept for the feasibility and therapeutic efficacy of the intrapulmonary transplantation of iPSC-derived macrophages in the context of herPAP. However, translation of this concept into a clinical setting, will require the genetic correction and functional repair of autologous iPSC-derived macrophages from patients with herPAP, as we already have demonstrated in previous work employing lentiviral as well as nuclease technology (19, 49). Moreover, clinical translation will require additional stringent and long-term safety testing in relevant animal models. Given the limitations of the humanized PAP model discussed above, we suggest that long-term safety studies be performed in an entirely murine model of PAP, such as Csf2Rb-deficient mice, as an essential prerequisite for clinical studies (19, 20). Specifically, it will be important to exclude teratoma formation as well as relevant immune reactions. An important safety feature of our approach, particularly regarding the risk of teratoma formation, is our transplantation of highly pure, differentiated macrophages. In our work, transplanted cells revealed a homogeneous macrophage phenotype and were entirely negative for pluripotency markers. On careful analysis, we did not observe any tumor formation in recipients. This is in accordance with previous work demonstrating a tumor-free outcome after transfer of iPSC-derived islet or hepatocyte-like cells in immunodeficient mice (50, 51). Another concern is potential immunological reactions to the therapeutic transgene after transplantation of gene-corrected autologous iPSC-derived macrophages. However, previous studies by us and others do not support this concern (19, 20).
In summary, we here introduce human iPSC–derived macrophages as an innovative source for pulmonary cell therapy. To the best of our knowledge, these data demonstrate for the first time the therapeutic potential of iPSC-derived cells in a pulmonary disease entity. Certainly, before clinical translation this approach requires careful studies regarding the long-term efficacy as well as safety of human iPSC–derived macrophages including their genetic modification. If successful, however, our data may be relevant beyond the rare disease of PAP and have profound implications for the treatment of other pulmonary diseases.
Acknowledgments
Acknowledgment
The authors thank Jana Bergmann, Christin Albrecht, Anika Dreier, Doreen Lüttge, and Theresa Buchegger (all of Hannover Medical School) for excellent technical assistance. In addition, the authors thank Matthias Ballmaier (Cell Sorting Facility, Hannover Medical School) for assistance in cell sorting, Heike Schneider (Research Core Unit Transcriptomics, Hannover Medical School) for preparation of libraries for RNA-Seq experiments, and Ewa Janosz (Institute of Experimental Hematology) for help with animal experiments.
Footnotes
Supported by grants from the German Center for Lung Research (DZL; G.H.), Deutsche Forschungsgemeinschaft (Cluster of Excellence REBIRTH; Exc 62/1 to T.M. and N.L., as well as DFG LA 3680/2-1), the Else Kröner-Fresenius-Stiftung (EKFS; 2015_A92 to N.L.; 2013_A24 to T.M. and G.H.), the Eva Luise and Horst Köhler Foundation for Rare Diseases (G.H., T.M., C.H., and N.L.), the Joachim Herz Stiftung (N.L.), the German Federal Ministry of Education and Research (CARPuD: 01GM0854, 01GM0857 to G.H.; iMACnet 01EK1602A to N.L., G.H., and T.M.) and MHH Hannover (HiLF grant, Hochschulinterne Leistungsförderung/Young Academy to C.H. and N.L.).
Author Contributions: G.H., C.H., N.L., and T.M. conceived the project and designed the experiments. C.H., M.A., G.G., K.T., T.G., T.S., C.C., S.G., O.D.-B., and B.T. conducted experiments or helped with analysis. A. Mucci and M.H. conducted experiments and helped with analysis. A. Mirenska conducted statistical analyses. G.H., C.H., N.L., and T.M. wrote the manuscript. All authors read and approved the manuscript.
Originally Published in Press as DOI: 10.1164/rccm.201708-1562OC on April 13, 2018
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 4.Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, et al. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol. 2012;30:1244–1248. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schöndorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun. 2014;5:4028. doi: 10.1038/ncomms5028. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, Mayhew C, Sallese A, Chalk C, Carey BC, Malik P, et al. Use of induced pluripotent stem cells to recapitulate pulmonary alveolar proteinosis pathogenesis. Am J Respir Crit Care Med. 2014;189:183–193. doi: 10.1164/rccm.201306-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Z, Zhang Y, Wang Y, Zhang D, Shen B, Luo M, et al. Progress of stem/progenitor cell–based therapy for retinal degeneration. J Transl Med. 2017;15:99. doi: 10.1186/s12967-017-1183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins F, Kramer P, Jacob A, Driver I, Thomas DC, McCauley KB, et al. Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells. J Clin Invest. 2017;127:2277–2294. doi: 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, et al. In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Stem Cell Reports. 2018;10:101–119. doi: 10.1016/j.stemcr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Gotoh S, Korogi Y, Seki M, Konishi S, Ikeo S, et al. Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat Methods. 2017;14:1097–1106. doi: 10.1038/nmeth.4448. [DOI] [PubMed] [Google Scholar]
- 12.Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, et al. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports. 2015;4:282–296. doi: 10.1016/j.stemcr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litvack ML, Wigle TJ, Lee J, Wang J, Ackerley C, Grunebaum E, et al. Alveolar-like stem cell–derived Myb− macrophages promote recovery and survival in airway disease. Am J Respir Crit Care Med. 2016;193:1219–1229. doi: 10.1164/rccm.201509-1838OC. [DOI] [PubMed] [Google Scholar]
- 14.van Wilgenburg B, Browne C, Vowles J, Cowley SA. Efficient, long term production of monocyte-derived macrophages from human pluripotent stem cells under partly-defined and fully-defined conditions. PLoS One. 2013;8:e71098. doi: 10.1371/journal.pone.0071098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dirksen U, Nishinakamura R, Groneck P, Hattenhorst U, Nogee L, Murray R, et al. Human pulmonary alveolar proteinosis associated with a defect in GM-CSF/IL-3/IL-5 receptor common β chain expression. J Clin Invest. 1997;100:2211–2217. doi: 10.1172/JCI119758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Moczygemba M, Doan ML, Elidemir O, Fan LL, Cheung SW, Lei JT, et al. Pulmonary alveolar proteinosis caused by deletion of the GM-CSFRα gene in the X chromosome pseudoautosomal region 1. J Exp Med. 2008;205:2711–2716. doi: 10.1084/jem.20080759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki T, Sakagami T, Rubin BK, Nogee LM, Wood RE, Zimmerman SL, et al. Familial pulmonary alveolar proteinosis caused by mutations in CSF2RA. J Exp Med. 2008;205:2703–2710. doi: 10.1084/jem.20080990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki T, Sakagami T, Young LR, Carey BC, Wood RE, Luisetti M, et al. Hereditary pulmonary alveolar proteinosis: pathogenesis, presentation, diagnosis, and therapy. Am J Respir Crit Care Med. 2010;182:1292–1304. doi: 10.1164/rccm.201002-0271OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, et al. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Happle C, Lachmann N, Škuljec J, Wetzke M, Ackermann M, Brennig S, et al. Pulmonary transplantation of macrophage progenitors as effective and long-lasting therapy for hereditary pulmonary alveolar proteinosis. Sci Transl Med. 2014;6:250ra113. doi: 10.1126/scitranslmed.3009750. [DOI] [PubMed] [Google Scholar]
- 21.Willinger T, Rongvaux A, Takizawa H, Yancopoulos GD, Valenzuela DM, Murphy AJ, et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc Natl Acad Sci USA. 2011;108:2390–2395. doi: 10.1073/pnas.1019682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finney-Hayward TK, Popa MO, Bahra P, Li S, Poll CT, Gosling M, et al. Expression of transient receptor potential C6 channels in human lung macrophages. Am J Respir Cell Mol Biol. 2010;43:296–304. doi: 10.1165/rcmb.2008-0373OC. [DOI] [PubMed] [Google Scholar]
- 23.Lange K, Holm L, Vang Nielsen K, Hahn A, Hofmann W, Kreipe H, et al. Telomere shortening and chromosomal instability in myelodysplastic syndromes. Genes Chromosomes Cancer. 2010;49:260–269. doi: 10.1002/gcc.20737. [DOI] [PubMed] [Google Scholar]
- 24.Dijkstra D, Hennig C, Witte T, Hansen G. Basophils from humans with systemic lupus erythematosus do not express MHC-II. Nat Med. 2012;18:488–489, author reply 489–490. doi: 10.1038/nm.2663. [DOI] [PubMed] [Google Scholar]
- 25.Manz MG. Human-hemato-lymphoid-system mice: opportunities and challenges. Immunity. 2007;26:537–541. doi: 10.1016/j.immuni.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Lachmann N, Happle C, Ackermann M, Lüttge D, Wetzke M, Merkert S, et al. Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am J Respir Crit Care Med. 2014;189:167–182. doi: 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- 27.Kuo LJ, Yang LX.γ-H2AX: a novel biomarker for DNA double-strand breaks In Vivo 200822305–309. [PubMed] [Google Scholar]
- 28.Trapnell BC, Whitsett JA, Nakata K. Pulmonary alveolar proteinosis. N Engl J Med. 2003;349:2527–2539. doi: 10.1056/NEJMra023226. [DOI] [PubMed] [Google Scholar]
- 29.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator–activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 31.Nakatani-Okuda A, Ueda H, Kashiwamura S, Sekiyama A, Kubota A, Fujita Y, et al. Protection against bleomycin-induced lung injury by IL-18 in mice. Am J Physiol Lung Cell Mol Physiol. 2005;289:L280–L287. doi: 10.1152/ajplung.00380.2004. [DOI] [PubMed] [Google Scholar]
- 32.Wojcik AJ, Skaflen MD, Srinivasan S, Hedrick CC. A critical role for ABCG1 in macrophage inflammation and lung homeostasis. J Immunol. 2008;180:4273–4282. doi: 10.4049/jimmunol.180.6.4273. [DOI] [PubMed] [Google Scholar]
- 33.Kleff V, Sorg UR, Bury C, Suzuki T, Rattmann I, Jerabek-Willemsen M, et al. Gene therapy of beta-c-deficient pulmonary alveolar proteinosis (βc-PAP): studies in a murine in vivo model. Mol Ther. 2008;16:757–764. doi: 10.1038/mt.2008.7. [DOI] [PubMed] [Google Scholar]
- 34.Nishinakamura R, Wiler R, Dirksen U, Morikawa Y, Arai K, Miyajima A, et al. The pulmonary alveolar proteinosis in granulocyte macrophage colony-stimulating factor/interleukins 3/5 βc receptor–deficient mice is reversed by bone marrow transplantation. J Exp Med. 1996;183:2657–2662. doi: 10.1084/jem.183.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doerschuk CM. Pulmonary alveolar proteinosis and macrophage transplantation. N Engl J Med. 2015;372:1762–1764. doi: 10.1056/NEJMcibr1413035. [DOI] [PubMed] [Google Scholar]
- 36.Kotton DN, Rossant J. Modeling pulmonary alveolar proteinosis with induced pluripotent stem cells. Am J Respir Crit Care Med. 2014;189:124–126. doi: 10.1164/rccm.201312-2122ED. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Xue C, Shah R, Bermingham K, Hinkle CC, Li W, et al. Functional analysis and transcriptomic profiling of iPSC-derived macrophages and their application in modeling Mendelian disease. Circ Res. 2015;117:17–28. doi: 10.1161/CIRCRESAHA.117.305860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mass E, Ballesteros I, Farlik M, Halbritter F, Günther P, Crozet L, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:aaf4238. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, et al. c-Myb+ erythro-myeloid progenitor–derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 43.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bellner L, Marrazzo G, van Rooijen N, Dunn MW, Abraham NG, Schwartzman ML. Heme oxygenase-2 deletion impairs macrophage function: implication in wound healing. FASEB J. 2015;29:105–115. doi: 10.1096/fj.14-256503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong Y, Sasidharan N, Laheji F, Frosch M, Musolino P, Tanzi R, et al. Microglial dysfunction as a key pathological change in adrenomyeloneuropathy. Ann Neurol. 2017;82:813–827. doi: 10.1002/ana.25085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M. Gene therapy comes of age. Science. 2018;359:eaan4672. doi: 10.1126/science.aan4672. [DOI] [PubMed] [Google Scholar]
- 47.Riolobos L, Hirata RK, Turtle CJ, Wang PR, Gornalusse GG, Zavajlevski M, et al. HLA engineering of human pluripotent stem cells. Mol Ther. 2013;21:1232–1241. doi: 10.1038/mt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Börger AK, Eicke D, Wolf C, Gras C, Aufderbeck S, Schulze K, et al. Generation of HLA-universal iPSC-derived megakaryocytes and platelets for survival under refractoriness conditions. Mol Med. 2016;22:274–285. doi: 10.2119/molmed.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhn A, Ackermann M, Mussolino C, Cathomen T, Lachmann N, Moritz T. TALEN-mediated functional correction of human iPSC–derived macrophages in context of hereditary pulmonary alveolar proteinosis. Sci Rep. 2017;7:15195. doi: 10.1038/s41598-017-14566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El Khatib MM, Ohmine S, Jacobus EJ, Tonne JM, Morsy SG, Holditch SJ, et al. Tumor-free transplantation of patient-derived induced pluripotent stem cell progeny for customized islet regeneration. Stem Cells Transl Med. 2016;5:694–702. doi: 10.5966/sctm.2015-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55:1193–1203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]