Figure 3.
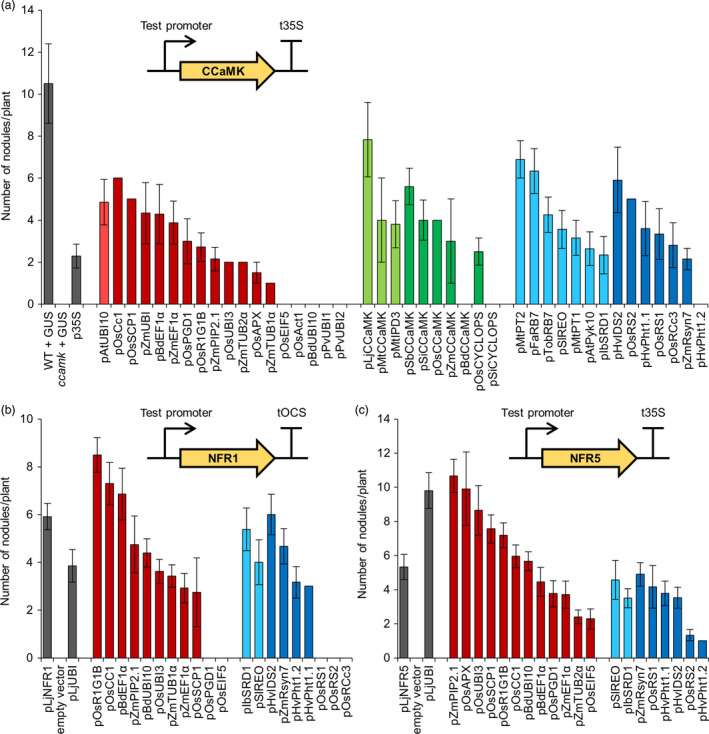
Different constitutive, symbiosis‐related and root‐specific promoters from the standard genetic parts library show varying abilities to complement the ccamk mutant of Medicago truncatula and the nfr1 and nfr5 mutants of Lotus japonicus. Constructs containing promoters from the standard genetic parts library driving expression of the CCaMK (a), NFR1 (b) or NFR5 (c) genes (see figure insets) were transformed into Medicago truncatula ccamk‐1 (a), Lotus japonicus nfr1‐1 (b) or nfr5‐2 (c) mutant plants via Agrobacterium rhizogenes‐mediated transformation. Total root nodule number was counted three weeks after inoculation with Sinorhizobium meliloti in M. truncatula, or five weeks after inoculation with Mesorhizobium loti in L. japonicus. Data represent mean ± standard error. In total, an average of at least 10 independently transformed plants was scored per construct (dsRED in the T‐DNA confirmed transformation). WT, wild type. Colour coding: control p35S and pLjUBI promoters, native pLjNFR1 and pLjNFR5 promoters, and control plants transformed with pAtUBI10‐GUS construct (GUS) or empty vector, grey; constitutive promoters, red; symbiosis‐related promoters, green; root‐specific promoters, blue; light and dark shades denote promoters from dicotyledonous and monocotyledonous species, respectively. Promoters pOsEIF5, pOsAct1, pBdUBI10, pPvUBI1, pPvUBI2, pBdCCaMK, pSiCYCLOPS and pHvPht1.2 showed no ccamk‐1 mutant complementation. Promoters pOsPGD1, pOsEIF5, pOsRS1, pOsRS2 and pOsRCc3 showed no nfr1‐1 mutant complementation.
