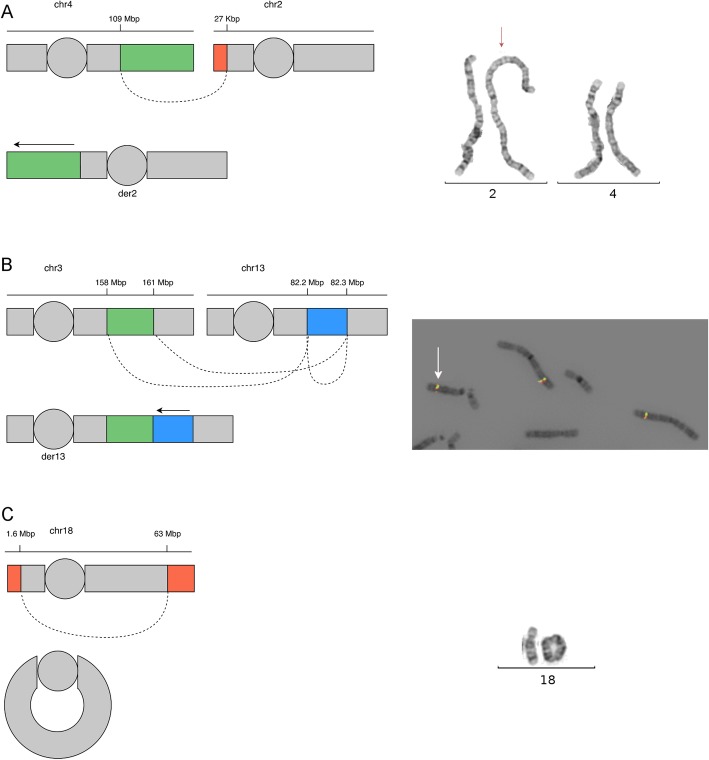
Fig. 3.
Three cases with complex genomic rearrangements resolved by WGS. a A schematic drawing of the 4q25q35.2 unbalanced translocation in individual RD_P406. The duplicated segment of 81 kb (green) is inserted into the p-arm of chromosome 2 directly before the telomeric sequences. A 27-kb deletion on chromosome 2 (orange) is visible in the WGS data. The dashed line represents the links from chromosome 4 to chromosome 2. To the right, the insertional duplication rearrangement is shown through karyotyping with the derivative chromosome 2 indicated by a red arrow. b A schematic drawing of the 3q25.32q26.1 insertional duplication in individual RD_P405 as in a. The duplicated segment of 2.23 Mb is inserted into chromosome 13, and a genomic segment of 69.6 kb on chromosome 13, adjacent to the insertion, has been inverted. To the right, FISH analysis using probes RP11-209H21SG (green) and RP11-203L15SO (red) located within the rearranged region on chromosome 3. In addition to two signals from chr 3q25.32q26.1, an extra signal is present on chromosome 13 (white arrow) verifying the location of the duplicated segment. c A schematic drawing of the r(18) present in individual RD_P414 as in a. To the right, the ring chromosome is shown through karyotyping