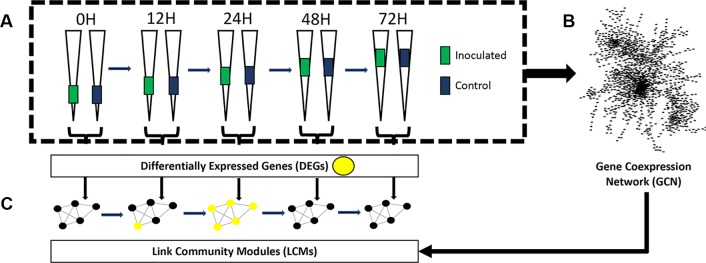
Figure 1.
Experimental overview. (A) Differentially expressed genes between control and inoculated samples were identified at each time point. Elongated triangles represent roots; colored portions are the areas harvested (see Methods) for each comparison in the bracket at the bottom. (B) A gene co-expression network was constructed from all 30 samples (see Methods) and (C) link community modules were identified by Knowledge Independent Network Construction. Differentially expressed link community modules (LCMs) were then identified by overlaying differentially expressed genes (yellow dots in modules) from each time point onto the LCMs.