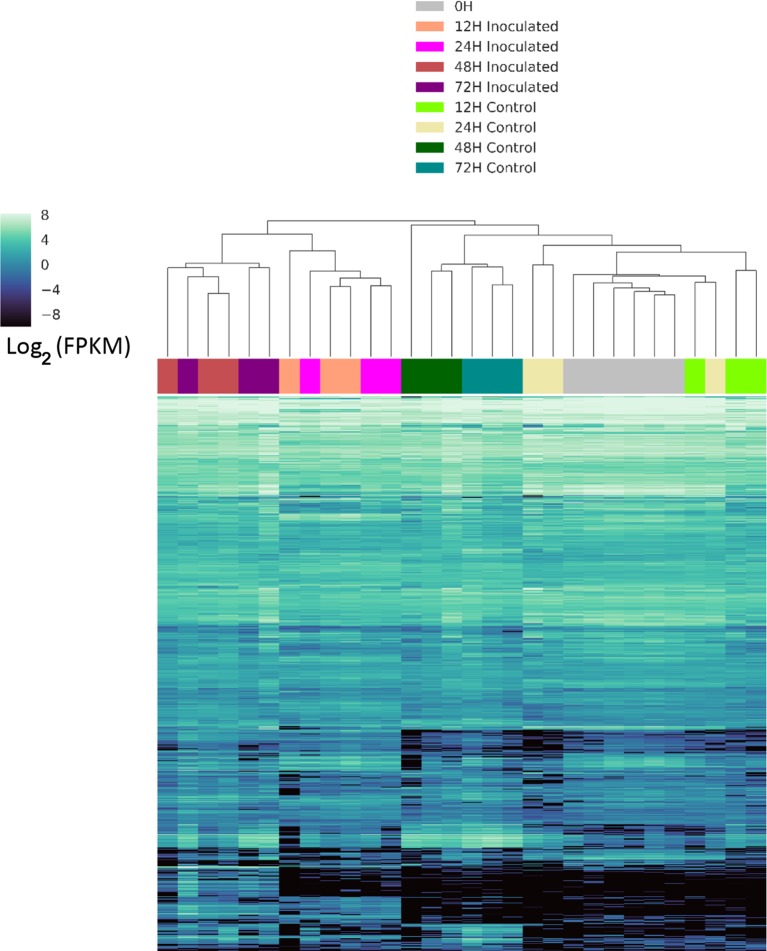
Figure 3.
Clustered heatmap of differentially expressed genes. Expression is reported as Log2 of the fragments per kilobase of gene per million read pairs. Samples were clustered and visualized using the Seaborn clustermap function, which uses Euclidian distance metrics to generate a linkage matrix used for hierarchical clustering. X-axis across the top is the is dendrogram of samples at individual time points and conditions, indicated by the color key, while the genes are represented by individual lines on the Y axis as sorted by the clustermap function.