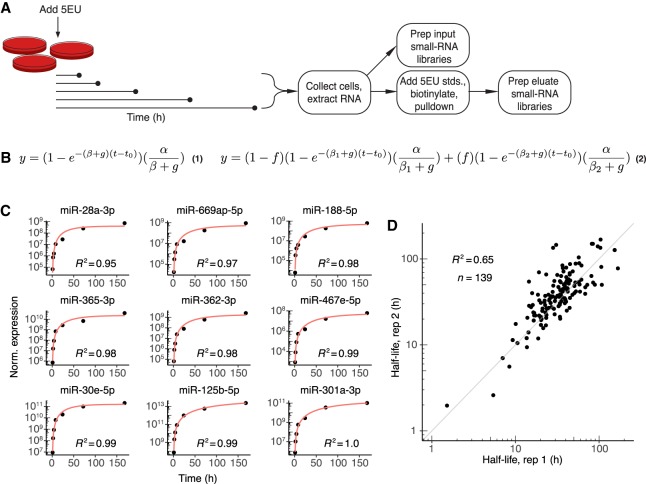
Figure 1.
Labeling with 5EU generates reproducible miRNA half-life values. (A) Schematic of the experimental setup. 5EU is added at time zero, and samples are collected at subsequent time intervals. RNA is extracted from each sample, and a small fraction of this extracted RNA is used to generate small-RNA sequencing libraries. The remainder of the RNA is mixed with quantitative standards (stds.), and molecules with one or more 5EU nucleotides are biotinylated and captured using streptavidin beads. RNA eluted from the beads is prepared for small-RNA sequencing. (B) Equations used for fitting approach-to-steady-state data. Equation 1 represents the single-exponential model used to fit a single decay rate for guide strands. Equation 2 represents the biexponential model used to fit two decay rates for both passenger and guide strands when assessing passenger-strand half-lives. Fits returned estimates for α (production rate), βn (degradation rate, with n distinguishing the two rates of Equation 2), f (fraction of molecules exhibiting the corresponding degradation rate), and t0 (time offset). The variable g describes the rate of cell doubling and was set manually based on experimental observations. Fits were carried out for each miRNA guide or passenger strand that satisfied an expression cut-off of 60 reads per million miRNA-mapping reads (i.e., 60 RPM) at each time point. (C) Representative fits to data from contact-inhibited MEFs. Data are plotted with black points, and single-exponential fits with a red line. Time courses representing the 10th through 90th deciles in goodness-of-fit are shown in order of increasing goodness-of-fit (from top left to bottom right). (D) Reproducibility of half-life measurements for guide strands. Shown is the relationship between values determined independently from each of the two biological replicates from contact-inhibited MEFs.