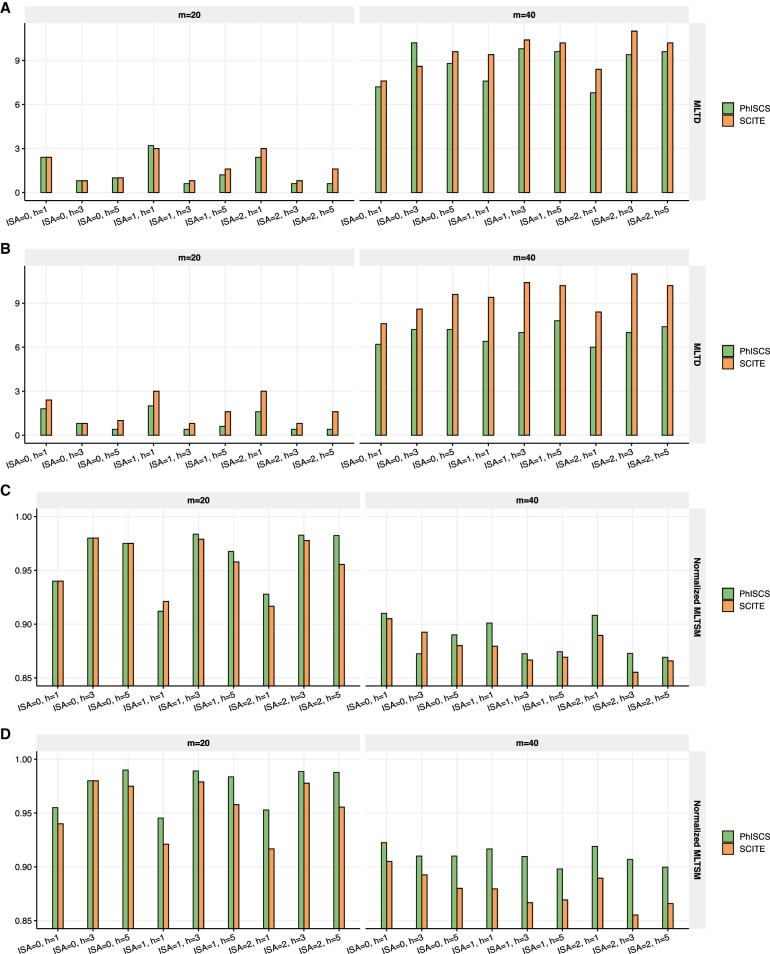
Figure 5.
Comparison of PhISCS with SCITE on simulated data where either one or two mutations are acquired at each node (in the latter case, one of the two mutations is an ISA violating mutation). Ten distinct trees of tumor evolution with either 20 mutations (i.e., s = 21, m = 20) or 40 mutations (i.e., s = 41, m = 40) were generated and 100 single cells sampled in each case. The false negative rate of single-cell data was set to 0.15. The depth of coverage for bulk data was set to 10,000×, and the minimum cellular prevalence of individual nodes was set to 3% for m = 20 and 2% for m = 40. A and C, respectively, show MLTD (dissimilarity) and normalized MLTSM (similarity) measure where ISA violations are allowed, but only SCS data was used as the input. B and D show, respectively, MLTD (dissimilarity) and normalized MLTSM (similarity) measure where ISA violations are allowed and both single-cell and bulk data are used as the input. On the x-axes, h, m, and ISA, respectively, denote the number of bulk samples, the total number of simulated mutations, and the number of simulated mutations for which ISA is violated.