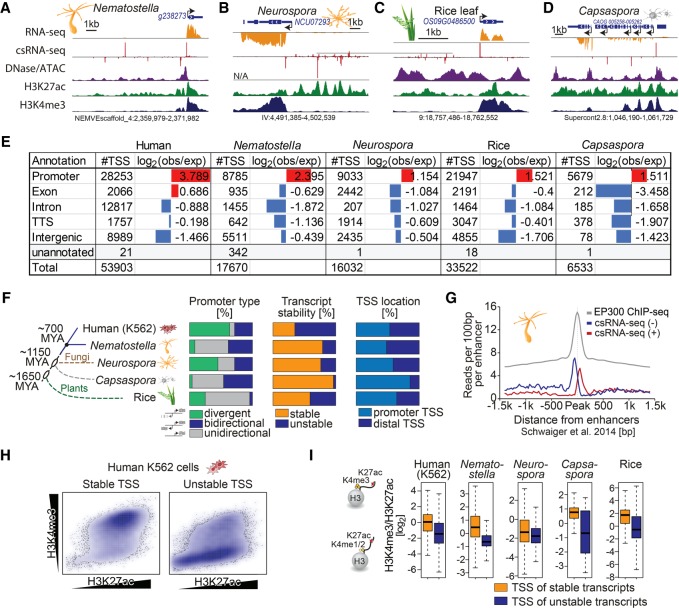
Figure 6.
csRNA-seq accurately profiles TSSs across eukaryotes. Capturing stable and unstable transcripts across eukaryotes by csRNA-seq reveals a gradual increase in unstable transcripts as more complex body plans evolved and conserved roles for the histone modifications H3K4me3 and H3K27ac. Example loci from diverse eukaryotes from across the kingdoms (A) Nematostella (metazoa), (B) Neurospora (fugi), (C) rice (plants), and (D) Capsaspora (protist). (E) Comparison of where TSSs defined by csRNA-seq are relative to genome annotations. (F) Species dendrogram with approximate divergence time and diagram of the percentage of stable versus unstable and unidirectional versus bidirectional transcripts. (G) csRNA-seq reads centered on the Nematostella enhancer regions defined by Schwaiger et al. (2014). (H) Scatterplot of H3K4me3 versus H3K27ac levels for stable and unstable transcripts from human K562 cells. (I) Boxplot with the log2 ratio of H3K27ac/H3K4me3 for the TSSs of stable and unstable transcripts.