The study of a family of alkaloid transporters in opium poppy shows a key role for alkaloid uptake in yeast strains engineered to produce opiate alkaloids.
Abstract
Although opiate biosynthesis has been largely elucidated, and cell-to-cell transport has been long postulated, benzylisoquinoline alkaloid (BIA) transporters from opium poppy (Papaver somniferum) have not been reported. Investigation of a purine permease-type sequence within a recently discovered opiate biosynthetic gene cluster led to the discovery of a family of nine homologs designated as BIA uptake permeases (BUPs). Initial expression studies in engineered yeast hosting segments of the opiate pathway showed that six of the nine BUP homologs facilitated dramatic increases in alkaloid yields. Closer examination revealed the ability to uptake a variety of BIAs and certain pathway precursors (e.g. dopamine), with each BUP displaying a unique substrate acceptance profile. Improvements in uptake for yeast expressing specific BUPs versus those devoid of the heterologous transporters were high for early intermediates (300- and 25-fold for dopamine and norcoclaurine, respectively), central pathway metabolites [10-fold for (S)-reticuline], and end products (30-fold for codeine). A coculture of three yeast strains, each harboring a different consecutive segment of the opiate pathway and BUP1, was able to convert exogenous Levodopa to 3 ± 4 mg/L codeine via a 14-step bioconversion process involving over a dozen enzymes. BUP1 is highly expressed in opium poppy latex and is localized to the plasma membrane. The discovery of the BUP transporter family expands the role of purine permease-type transporters in specialized metabolism, and provides key insight into the cellular mechanisms involved in opiate alkaloid biosynthesis in opium poppy.
Benzylisoquinoline alkaloids (BIAs) are a structurally diverse class of ∼2,500 specialized metabolites with a common phylogenetic and biosynthetic origin, beginning with the condensation of two Tyr derivatives, in the Ranunculales order of plants (Liscombe et al., 2005). Many of humanity’s most ancient medicines, poisons, hunting aids, and ceremonial preparations are derived from plants accumulating BIAs (Hagel et al., 2015). Opium poppy (Papaver somniferum) is a notable BIA producer and is the sole commercial supply of key pharmaceuticals such as codeine, morphine, and the precursor thebaine, needed for the semisynthesis of naloxone, an opioid-overdose antidote. Naloxone is not a cure for addiction, but like a defibrillator for a heart attack, it revives and creates the possibility for treatment and rehabilitation. Costs for various formulations of naloxone, fueled by demand, drug shortage, and opaque pricing, have at times increased from 244% to 3,797% (Rosenberg et al., 2018).
Reliance on opium poppy as a source material for opiates is problematic due to agronomic fluctuation exacerbated by factors such as climate change. On the other hand, total chemical synthesis of opiates is expensive and elusive due to complex stereochemistry. Indeed, even chemical derivatization of plant-derived thebaine is afflicted by requirements for harsh conditions and hazardous materials (e.g. cyanogen bromide and chloroformate; Reed and Hudlicky, 2015). Metabolic engineering of microbes with alkaloid biosynthetic machinery from opium poppy can circumvent these challenges and provide a sustainable, scalable, and dynamic resource for opiate pharmaceuticals. De novo thebaine biosynthesis has been achieved in Saccharomyces cerevisiae, although at very low yields (∼6 μg/L culture thebaine; Galanie et al., 2015). Titers are continually improving through the discovery of new enzymes (Chen et al., 2018; Dastmalchi et al., 2019). Yet there remains a remarkable dearth in knowledge regarding how these products are transported within the plant, a problem which has conceivably limited production in alternative, microbial based systems.
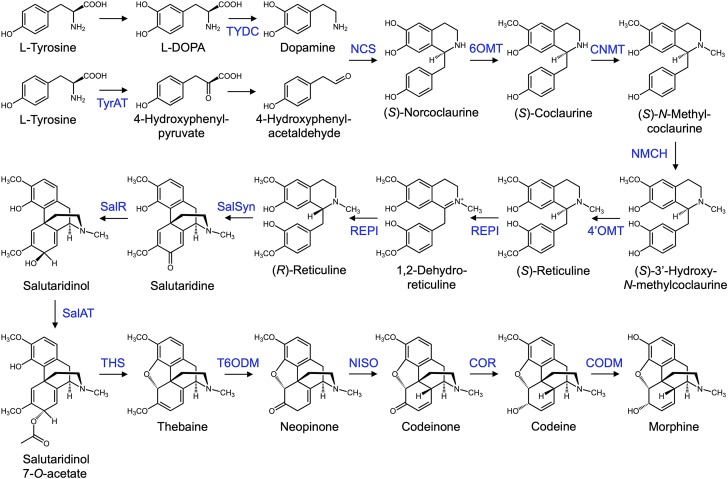
BIA metabolism in opium poppy begins with a core set of reactions leading to the central metabolite (S)-reticuline (Fig. 1). While the sequence of initial reactions leading from Tyr to dopamine and aldehyde precursors are not fully resolved in planta, steps likely include hydroxylation, decarboxylation, and transamination (Lee and Facchini, 2011). Recent efforts by microbial engineers to reconstitute BIA metabolism in Escherichia coli (Nakagawa et al., 2016) and yeast (Galanie et al., 2015; Chen et al., 2018) have sidestepped this uncertainty by incorporating a bacterial enzyme (dihydroxyphenylalanine [DOPA] decarboxylase [DODC]) that decarboxylates Levodopa (l-DOPA), provided in the culture media, to dopamine.
Figure 1.
Opiate biosynthetic pathway occurring in opium poppy beginning with two molecules of Tyr and ending with morphine. Initial portions of the pathway are shared with papaverine and noscapine biosynthetic routes (not shown), which bifurcate at (S)-coclaurine and (S)-reticuline, respectively. Enzyme catalysts are indicated above reaction arrows. Additional abbreviations: 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine N-methyltransferase; COR, codeinone reductase; NISO, neopinone isomerase; TyrAT, tyrosine aminotransferase; TYDC, tyrosine decarboxylase.
A second modification has been engineered in the pathway leading to (S)-reticuline to avoid a perceived bottleneck in the activity of a cytochrome P450 (CYP), N-methylcoclaurine 3′-hydroxylase. In the plant, with the aid of norcoclaurine synthase (NCS), dopamine and 4-hydroxyphenylacetaldehyde (4-HPAA) condense to form (S)-norcoclaurine (NCC). This is followed by four reactions catalyzed by enzymes including N-methylcoclaurine 3′-hydroxylase. In engineered microbes, NCS combines dopamine with 3,4-dihydroxyphenylacetaldehyde in place of 4-HPAA to form (S)-norlaudanosoline (NLDS). NLDS is catalyzed by three reactions to yield (S)-reticuline, thereby reducing the pathway by one step and omitting the inefficient CYP activity. To build this alternative NLDS-dependent route, the human enzyme monoamine oxidase A (MAO-A) is included to biosynthesize dopamine with 3,4-dihydroxyphenylacetaldehyde.
Formation of (S)-reticuline is followed by the action of several enzymes to yield the first opiate, thebaine: (S)-reticuline epimerase (REPI), salutaridine synthase (SalSyn), salutaridine reductase (SalR), salutaridinol 7-O-acetyltransferase (SalAT), and the recently discovered thebaine synthase (THS; Chen et al., 2018). A cytochrome P450 reductase enzyme is added as a reductase partner to SalSyn, a CYP enzyme. Finally, thebaine is converted to morphine by thebaine 6-O-demethylase (T6ODM), codeinone reductase, and codeine O-demethylase (CODM).
The pathway described above, starting with the condensation of NCC and ultimately forming morphine, requires the participation of three distinct cell types (Bird et al., 2003; Onoyovwe et al., 2013). Many of the enzymes are synthesized in companion cells, while the activity of early- and latter-stage opiate metabolism is largely partitioned into sieve element and laticifer cells, respectively. The end products codeine and morphine are accumulated in the cytoplasm of laticifers, the alkaloid-rich latex (Roberts et al., 1983; Weid et al., 2004; Onoyovwe et al., 2013). In this segmented pathway, intermediates are presumably translocated from sieve elements to laticifers. However, the mechanisms of opiate transport have not been characterized. The antimicrobial berberine is one of the few BIAs with a characterized mechanism of transport. Berberine is also derived from (S)-reticuline and has been studied in Japanese goldthread (Coptis japonica). Current models suggest that transporters from the ATP-binding cassette (ABC) family mediate the translocation of berberine from root to rhizome tissues, across the plasma membrane (Shitan et al., 2003, 2013). There, a multiantimicrobial extrusion protein (MATE) family transporter, localized to the tonoplast, imports berberine into the vacuole (Takanashi et al., 2017). Neither ABC nor MATE proteins have been implicated in opiate biosynthesis.
A family of transporters with known affinity for alkaloids are purine uptake permeases (PUPs), originally described in Arabidopsis (Arabidopsis thaliana; Gillissen et al., 2000). PUPs transport a variety of nitrogenous substrates and derivatives that are primarily purine nucleobase, such as adenine and cytokinin hormones, but they have expanded activities with nonpurine alkaloids such as vitamin B6 (Jelesko, 2012; Szydlowski et al., 2013; Kato et al., 2015). Well-known purine specialized metabolites are two plant alkaloids, caffeine and theobromine, which competitively bind to PUPs. The role of PUPs was further expanded when a homolog (nicotine uptake permease 1 [NtNUP1]) was implicated in nicotine transport in tobacco (Nicotiana tabacum; Hildreth et al., 2011). Indeed, nicotine is not a purine-derived alkaloid and the activity of NtNUP1 enlarges the catalog of possible substrates for PUP-type transporters. NtNUP1 is plasma membrane localized and could play a role in the movement of nicotine from its point of synthesis in the roots to aerial organs, where the MATE transporter JAT1 mediates vacuolar accumulation (Morita et al., 2009).
As part of our ongoing investigations regarding BIA biosynthesis, we assembled a draft model of the opium poppy genome and noticed a PUP-like sequence within a cluster of genes encoding several opiate biosynthetic enzymes (Chen et al., 2018; Facchini et al., 2018). Using this sequence as a query, we identified nine PUP-like homologs in transcriptomes of opium poppy, several of which function as BIA transporters. Herein, we describe the discovery, characterization, and deployment in a synthetic biology context of a distinct subfamily of PUP homologs designated as BIA uptake permeases (BUPs).
RESULTS
Putative PUP Clustered with Thebaine Pathway Genes
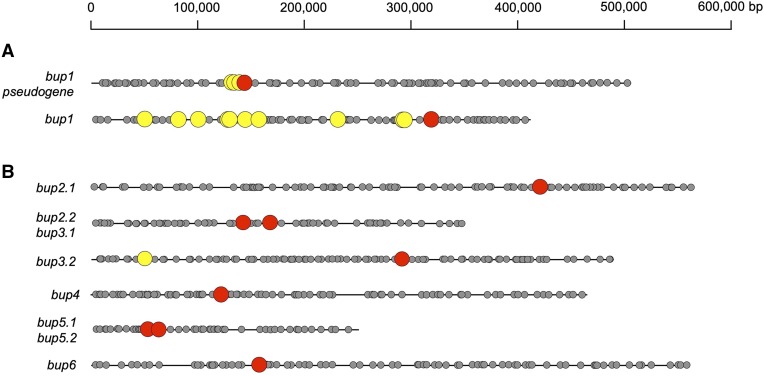
In a previous article, a draft genome for the opium poppy chemotype Roxanne revealed the clustering of five genes involved in the biosynthesis of thebaine from (S)-reticuline (Chen et al., 2018), including those encoding SalR, SalAT, and THS. This cluster was investigated for other genes that might encode metabolic, accessory, or transport proteins. Within this cluster were two regions annotated as encoding putative PUPs, which appeared to be identical although sequence quality was insufficient to enable confident analysis. We recently completed an improved draft of the opium poppy genome (Dastmalchi et al., 2019), enabling a more detailed analysis, which revealed only a single copy of the putative PUP gene linked to SalR, SalAT, and THS genes (Fig. 2A; Supplemental Fig. S1). The genomic scaffold containing this cluster is ∼500,000 bp, although the region containing the tightly linked SalR, SalAT, THS, and putative PUP genes extends only from ∼134,000 to 152,000 bp. The putative PUP encoding region is present in antisense orientation compared with nearby SalR and THS, and mapping with available transcriptome resources revealed the presence of two exons (151,229–150,459 and 150,337–149,973 bp for exons 1 and 2, respectively). There are single-base pair insertions in both exon 1 and exon 2 (at positions 151,130 and 150,100, respectively) that render potential frameshifts and would encode premature “stop” codons in the cognate protein. Therefore, this genetic sequence was annotated as a pseudogene and is referred to hereafter as BUP1 pseudogene, named as such due to its proximity to BIA pathway genes. BUP1 pseudogene was used to query extensive opium poppy transcriptomic resources (Desgagné-Penix et al., 2010), revealing an apparent BUP1 complementary DNA (cDNA) free of frameshifts or premature stop codons but otherwise mapping with 100% identity to the BUP1 pseudogene (Supplemental Datasets S1 and S2). The presence of this transcript suggested that at least one functional copy of BUP1 existed in the opium poppy genome. Indeed, mapping the genome with BUP1 revealed a second, apparently functional copy free of mutation within a shorter scaffold (∼400,000 bp; Fig. 2A) containing a more diffusely arranged set of BIA biosynthesizing genes. These linked genes encoded several enzymes within noscapine metabolism, including noscapine synthase, 3-O-acetylpapaveroxine carboxylesterase, 1,13-dihydroxy-N-methylcanadine 13-O-acetyltransferase, 1-hydroxy-13-O-acetyl-N-methylcanadine 8-hydroxylase, and 1-hydroxy-N-methylcanadine 13-hydroxylase (Winzer et al., 2012; Dang et al., 2015; Supplemental Fig. S2; Supplemental Dataset S3), thereby suggesting that the scaffold represented a previously reported “noscapine” genomic cluster (Winzer et al., 2012). As this gene was also clustered with BIA biosynthesizing genes, but potentially functional, it was named BUP1.
Figure 2.
Occurrence of predicted BUP-encoding genes (red) on eight different opium poppy genomic scaffolds. A, Genomic organization of bup1 and a bup1 pseudogene. B and C, Genomic environments of other functional bup genes. Genes encoding enzymes with known or purported roles in BIA metabolism (yellow) formed diffuse (scf7180001505306) and tight (scf7180001505888) clusters with BUP1 and a truncated BUP1 pseudogene, respectively. Other BUPs, which did not appear clustered with BIA biosynthetic genes, were linked with other BUPs or were found alone. An exception was BUP3.2, which was found ∼0.24 Mbp from a gene putatively encoding CODM (scf7180001507141). Gray circles represent other putative genes predicted using the Augustus feature of Geneious 11.1.2, using Arabidopsis tool settings (Stanke et al., 2004).
BUP Homologs Improve Modular Bioconversion of BIA Substrates in Yeast
Using the BUP1 transcript as a query, further mining of transcriptomes to a cutoff of 40% sequence identity revealed eight additional BUPs (BUP2–BUP9). Phylogenetic relationships of opium poppy BUPs (deduced amino acids) were analyzed together with homologs from other plant species, including PUPs from Arabidopsis and rice and NtNUP1 from tobacco (Supplemental Fig. S3). BUP1 through BUP8 form a tight opium poppy-specific clade. BUP9 forms a branch with additional predicted opium PUPs with very low sequence identity (<30%) to BUP1, labeled transport proteins. Multiple amino acid sequence alignment revealed conserved regions across all PUP sequences, spanning both opium poppy and other plant species (Supplemental Fig. S4). As an initial screen for functionality, we routinely employ “plug-and-play”-based evaluation of new gene candidates in engineered yeast (Morris et al., 2016). Briefly, this methodology involves plasmid-based expression of new gene candidates within the context of S. cerevisiae strains already capable of biosynthesizing BIA products from simple precursors (e.g. DOPA, dopamine, and NLDS) or pathway intermediates (reticuline, thebaine, and codeine) owing to genomic integration of multiple pathway genes. This method circumvents problems of limited substrate availability (e.g. rare or expensive alkaloids), exposes new candidates to a multitude of potential substrates at once, and may reveal an impact on pathway functionality (e.g. increased intermediate/product levels). Gene candidates whose expression in engineered microbes causes perturbations in alkaloid levels are then explored in more detail to ascertain specific functions. This plug-and-play approach has been used repeatedly as part of our gene discovery pipeline (Chang et al., 2015; Chen et al., 2018; Dastmalchi et al., 2019) and is applied herein for BUP screening. We sought to explore potential functionality of BUP genes by expressing their cDNA individually in three different engineered yeast strains, supporting three “blocks” of opiate metabolism, respectively (Supplemental Fig. S5). The three yeast strains genomically engineered for metabolite conversion assays (Supplemental Fig. S5C) included the metabolic components derived from opium poppy (natural and synthetic variants), Pseudomonas putida (DODC), and Homo sapiens (MAO-A) for the transformation of (1) l-DOPA to (S)-reticuline; (2) (S)-reticuline to thebaine; and (3) thebaine to morphine. Each strain was transformed separately with a BUP (BUP1–BUP9) transient expression plasmid and cultured with substrate supplementation as indicated (Fig. 3; Supplemental Fig. S5, A and C). The “empty vector” control in these assays is the genomically engineered yeast strain (1, 2, or 3) carrying the requisite metabolic components in addition to a plasmid without any BUP cDNA. We restricted the monitoring of conversion products to media rather than, or in addition to, cellular lysates, as relatively little alkaloid is retained in cells. This phenomenon is demonstrated in Supplemental Fig. S6, which compares reticuline product levels in strain 1 media and lysates following batch-feeding of NLDS. (S)-Reticuline is taken up with relative efficiency by most BUPs (Fig. 4), yet most remains in the media following bioconversion; for example, >1% of total reticuline is retained in cells for BUP4, up to a maximum of 11.3% for BUP2 (Supplemental Fig. S6). It is noteworthy that yeast cultures harboring BUP4 and BUP5 appear capable of turning over (S)-reticuline product(s). This result was unexpected, as neither BUP4 nor BUP5 showed significant capacity for NLDS uptake in wild-type yeast transformed with BUP-expressing plasmids (Fig. 4). Closer examination of (S)-reticuline of the strain 1 culture medium (Supplemental Fig. S6) showed that cells expressing BUP4 and BUP5 exhibit the lowest reticuline levels of all tested cultures. One possible explanation for the apparent incongruity of these results is that NLDS uptake by BUP4 and BUP5 is minimal, but not entirely negligible, allowing some conversion through engineered pathways to a small pool of reticuline in spite of steady-state intracellular levels that are generally below detection limits.
Figure 3.
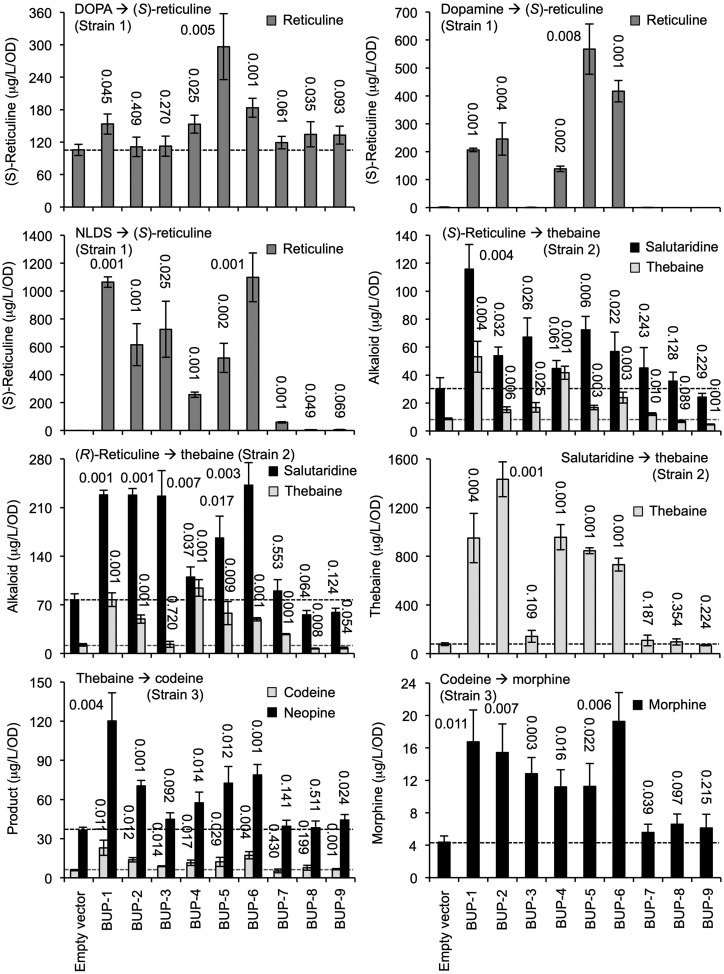
Metabolic conversion assay results. Engineered S. cerevisiae strains 1, 2, or 3 were transformed with one of nine plasmids, allowing expression of BUP1–BUP9, respectively (x axis). Yeast were exogenously fed the substrate/intermediate indicated, and media were monitored postincubation for the metabolite(s) shown. For comparison, dotted lines indicate levels of metabolite product detectable in control assays lacking BUP expression. The se was calculated using four biological replicates representing four individual transformants. The number above each bar indicates the P value obtained using an unpaired, two-tailed Student’s t test to compare with the empty vector control.
Figure 4.
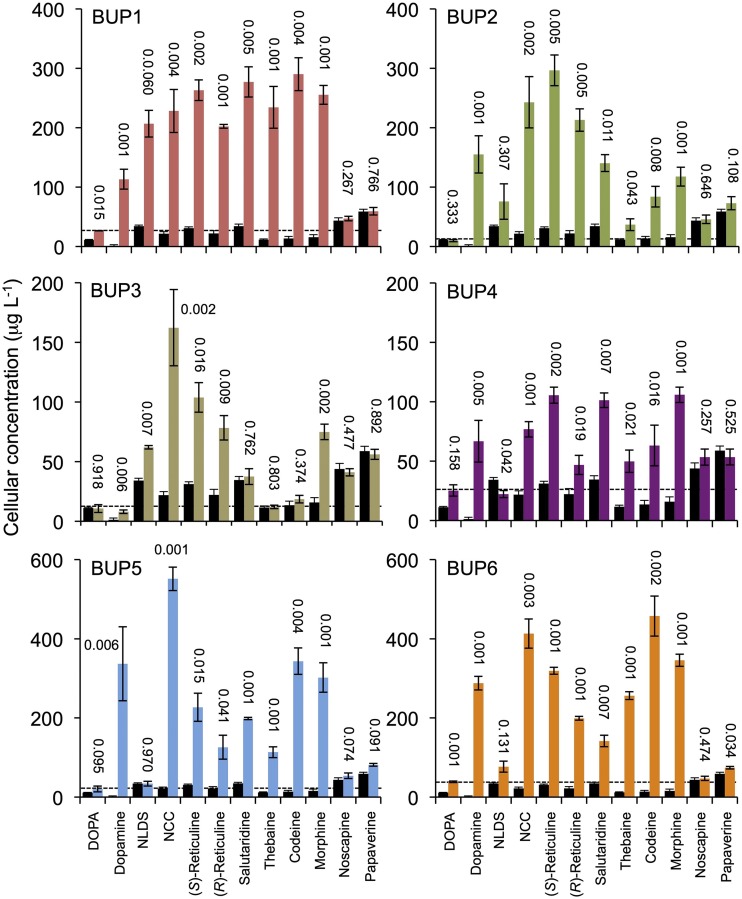
Uptake assay results. S. cerevisiae was transformed with one of six plasmids, allowing expression of BUP1–BUP6, respectively. Substrates provided exogenously in the media and measured postincubation in cellular extract are indicated on the x axes. Black bars and colored bars represent empty vector and BUP-expressing strains, respectively. The se was calculated using four biological replicates representing four individual transformants. The number above each bar indicates the P value obtained using an unpaired, two-tailed Student’s t test to compare with the empty vector control.
Strain 1 cultures were fed with the starting-point metabolite, l-DOPA, or intermediates dopamine or NLDS (Fig. 3). The yield of final product, (S)-reticuline, was measured in comparison to the empty vector. Cultures carrying BUP5 and BUP6 showed the largest significant improvement in (S)-reticuline production when fed l-DOPA or dopamine. BUP1- and BUP6-expressing cultures produced the highest levels of (S)-reticuline when fed NLDS. Titers were doubled in the BUP6 line when l-DOPA was replaced with dopamine (183 ± 28 to 416 ± 74 μg L−1 optical density [OD]−1) and further increased to 1,099 ± 395 μg L−1 OD−1 when NLDS was supplemented. Highest overall titers were recorded when the intermediate NLDS was provided to the engineered yeast cultures. Expression of BUP7, BUP8, or BUP9 had marginal or no significant impact on strain 1 production of (S)-reticuline, irrespective of the substrate supplemented.
Strain 2 cultures were either supplemented with (S)- or (R)-reticuline recording salutaridine and thebaine, or were fed salutaridine and recording thebaine production (Fig. 3). BUP1, BUP2, BUP3, BUP5, and BUP6 expression improved the production of the intermediate salutaridine, but only BUP1, BUP4, and BUP6 improved thebaine levels. BUP4 did not significantly affect intermediate production but revealed a positive impact on the end product, thebaine. Providing (R)-reticuline effectively bypassed the first enzymatic step, catalyzed by REPI, yielding larger titers of salutaridine and thebaine for all strain 2 cultures, including the empty-vector control. Expression of BUP1–BUP3, BUP5, and BUP6 improved salutaridine, and the same cultures, except for BUP3, also improved thebaine production. The presence of plasmids carrying BUP7, BUP8, or BUP9 did not affect production of the intermediate or the end product.
Supplementation of the midpoint intermediate salutaridine improved titers 6- to 9-fold for the empty vector control and reached a maximum output of 1,433 ± 321 μg L−1 OD−1 of thebaine with the transient expression of BUP2, followed by BUP1 and BUP4–BUP6, in descending order of impact. As in previous assays with strain 2, the addition of BUP7, BUP8, or BUP9 did not positively affect the metabolic output of this pathway module. Additionally, BUP3 did not affect thebaine titers, consistent with the previous two assays, where it only improved salutaridine levels.
Finally, strain 3 cultures were either fed thebaine, recording levels of codeine, and the isomeric byproduct neopine or were supplemented with codeine while recording morphine titers (Fig. 3). Significant improvement in both codeine and neopine production was observed for BUP1, BUP2, and BUP6. Codeine-supplemented cultures produced larger titers of morphine with the addition of any BUP expression plasmid except BUP7–BUP9. The highest yields of morphine were achieved with BUP6, BUP1, and BUP2, in descending order. Characterization of the BUP transporter family revealed that BUP1 was consistently effective in facilitating opiate production from l-DOPA, (S)-reticuline, and thebaine in the three modular strains, improving titers of intermediates and end products for each strain.
Genome Linkage Analysis for BUP Genes with Potential Roles in Alkaloid Transport
Initial screening of BUP functionality through plug-and-play methodology revealed BUP1–BUP6 as potentially involved in alkaloid transport, while no potential impact was observed for BUP7–BUP9. As it was known that BUP1 maps to clustered regions of BIA biosynthetic genes, we sought to investigate whether this was true for other BUPs with potential roles in alkaloid metabolism. Transcripts for active BUPs (BUP2–BUP6) were mapped onto eight different genomic scaffolds (Fig. 2B; Supplemental Figs. S7–S12; Supplemental Dataset S3). Clustering of BUP3.2 (the second copy of BUP3) and CODM within a 0.24 Mbp region was identified (Fig. 2B). This BUP3.2-containing scaffold also hosted a copy of BUP9 (i.e. BUP9.2; Supplemental Fig. S9). The remaining BUPs are situated on scaffolds either individually or in proximity to BUP putative paralogs (Supplemental Figs. S7–S12). Two copies of each of BUP2, BUP3, and BUP5 were identified on the opium poppy genomic scaffolds, in addition to two further BUP3-like genes.
BUPs Are Uptake Transporters of BIA Metabolites
To gain insight regarding specific transport abilities, BUP1–BUP6 were subjected to cellular uptake assays. BUP7–BU9 were not part of this study, as metabolic conversion assays indicated that they had no impact on alkaloid metabolism. The uptake assays were conducted by exogenous supplementation of substrates (l-DOPA, dopamine, or BIAs) to yeast strains expressing heterologous BUP cDNA and recording of cellular concentrations of substrates 24 h after feeding (Supplemental Fig. S5B). A 24-h incubation period was chosen based on preliminary time-course screens of S. cerevisiae expressing BUP1, where it was found that concentrations of exogenously fed substrate were approaching maximum levels, or had reached maximum levels, in cell extracts (Supplemental Fig. S13). The substrate uptake assays with BUP1–BUP6 were evaluated against an empty vector control to account for the potential activity of endogenous transporters or other modes of yeast uptake.
BUP1 significantly increased the cellular concentration of yeast with all substrates except noscapine and papaverine (Fig. 4). The most dramatic rise in cellular uptake occurs for substrates from dopamine to morphine, representing intermediates forming the first committed step of BIA metabolism (dopamine and 4-HPAA condensing to form NCC) to the canonical end product (morphine). A marginal, yet significant, increase was recorded for the uptake of l-DOPA, which directly precedes dopamine in the pathway. Except for a minor (P = 0.034) increase in cellular concentrations of papaverine for BUP6, the lack of uptake potential for noscapine and papaverine was recorded for all six BUPs.
The pattern of cellular uptake displayed by BUP1-expressing yeast, the increased uptake of metabolites from dopamine to morphine, and the lack of improvement for l-DOPA, noscapine, or papaverine uptake, is largely conserved by BUP2–BUP6, with some exceptions. BUP2 has lower, yet significant, activity with thebaine, codeine, and morphine, relative to other metabolites. BUP3 does not appear to significantly increase salutaridine, thebaine, or codeine uptake as compared to the empty vector control. BUP4–BUP6 do not significantly improve NLDS uptake. BUP5 facilitates the largest (∼300×) fold increase in dopamine uptake.
BUP1 Facilitates Modular Coculturing of BIA Biosynthesis
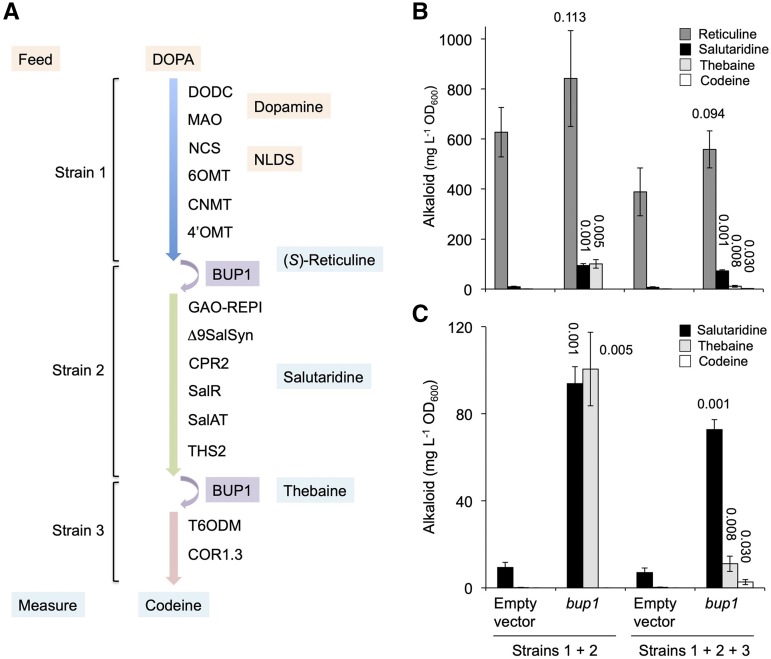
The ability of BUPs to import alkaloids rendered them potentially important tools for synthetic biology purposes. It is well known that establishing long, plant-derived, biosynthetic pathways in microorganisms for the ultimate goal of developing fermentation-based, alternative production systems for valuable pharmaceuticals is plagued with loss of pathway intermediates to the surrounding media, leading to poor yields of end products (Galanie et al., 2015). The yeast-based endogenous mechanism(s) underpinning intermediate loss are unknown, although reimport of pathway metabolites by transporters such as BUPs could represent a means to increase product yields. While a yeast strain hosting a “complete” alkaloid biosynthetic pathway from l-DOPA to opiates was not available for this study, coculturing of three engineered yeast strains, each expressing a BUP1 plasmid, would theoretically enable complete conversion from l-DOPA to codeine. BUP1, which was chosen for this study owing to its import ability with a broad suite of alkaloids (Fig. 4), was expected to act as a “bridge” between strains by importing key intermediates (Fig. 5). The metabolic output was compared to cocultured strains transformed with plasmid devoid of BUP1 (empty vector). Slight changes were made to the strains used in this study compared with those used for plug-and-play (metabolic conversion) assays (Fig. 3; Supplemental Fig. S5C). These changes included replacing germacrene A oxidase (GAO)-REPI and SalSynΔ9 with REPI-NT59 and SalSyn-NT137, respectively, and truncating the third module to exclude CODM. Cocultured strains were supplemented with l-DOPA and the concentrations of the alkaloids (S/R)-reticuline, salutaridine, thebaine, and codeine were recorded (Fig. 5). Coculturing included either a “binary” (1 + 2) or “tertiary” (1 + 2 + 3) combination, in conjunction with the absence or presence of BUP1.
Figure 5.
Modular coculturing of S. cerevisiae strains expressing BUP1 facilitates opiate production from l-DOPA. A, Each strain, transformed with a plasmid for BUP1 expression, was genomically integrated with the BIA biosynthetic genes indicated. Strains 1–3 were engineered to produce (S)-reticuline, thebaine, and codeine, respectively. “Binary” (strains 1 + 2) or “tertiary” (1 + 2 + 3) cocultures were fed l-DOPA and incubated for 24 h, after which media were profiled for (S)-reticuline, salutaridine, thebaine, and codeine. B and C, The same data but with differently scaled y axes to highlight quantitative data variations. 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine N-methyltransferase; 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; CPR, cytochrome P450 reductase; COR, codeinone reductase. The se was calculated using four biological replicates representing four individual transformants. The number above each bar indicates the P value obtained using an unpaired, two-tailed Student’s t test to compare with appropriate empty vector controls.
Cocultured yeast supplemented with l-DOPA, including the empty vector controls, formed (S)-reticuline as a result of the activity of the “strain 1” module enzymes. The expression of BUP1 in the cocultures significantly improved levels of the downstream intermediates salutaridine and thebaine. This result confirmed that BUP1 acted to import (S)-reticuline, which had been exported and/or lost to the surrounding media by strain 1, to strain 2 for further conversion. In the binary coculture, BUP1 expression significantly improved salutaridine titers (10.1-fold) and led to an equivalent accumulation of thebaine (101 ± 38 mg L−1 OD−1). The addition of strain 3 led to an overall reduction in metabolic output. However, closer analysis revealed that BUP1-expressing tertiary cultures produce the desired end product, codeine (3 ± 4 mg L−1 OD−1), via the activity of the third-strain enzymes (T6ODM and codeinone reductase). Codeine was absent from the tertiary cocultured yeast lacking BUP1 expression.
BUP1 Is a Laticifer-Specific Plasma Membrane Transporter
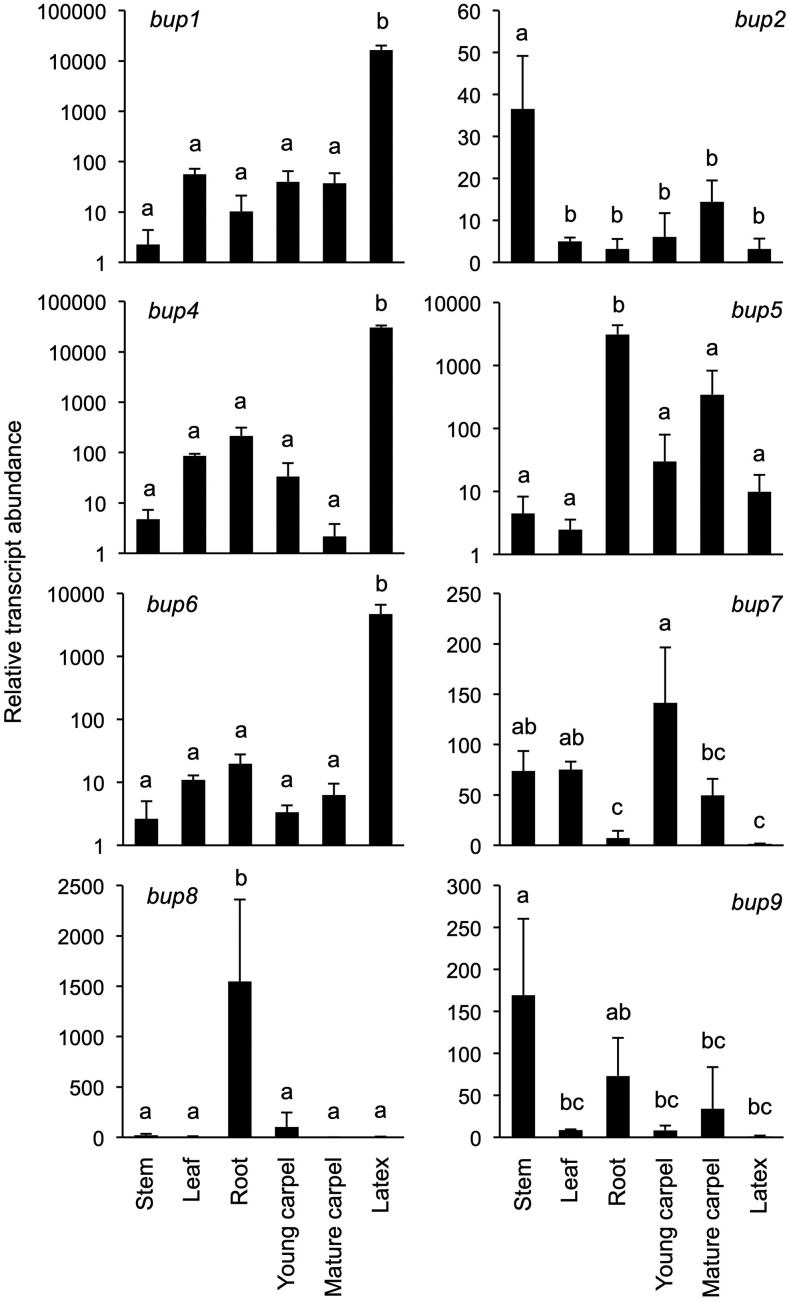
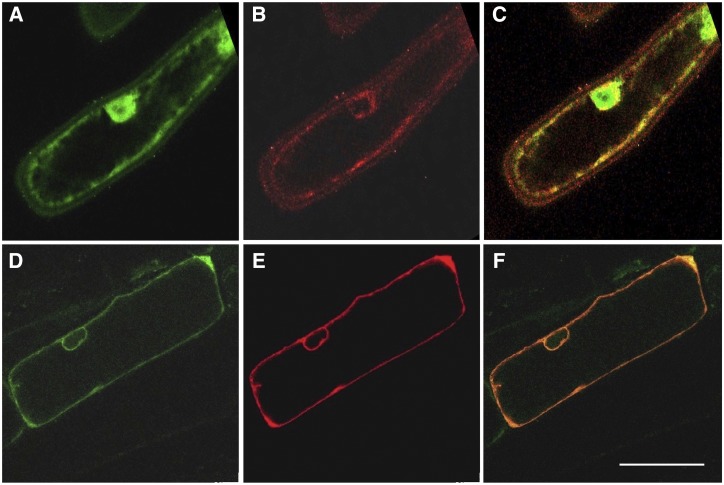
With roles in alkaloid transport established for heterologous yeast systems, we turned our attention to putative functional relevance in planta. Relative expression of BUP genes in the opium poppy chemotype Roxanne was measured by reverse-transcription quantitative PCR (RT-qPCR; Fig. 6). BUP1, BUP4, and BUP6 share a similar pattern of organ-specific gene expression, as they are all predominantly expressed in the latex. It is noteworthy that amino acids for these BUPs form a monophyletic cluster, together with BUP7 (Supplemental Fig. S3). BUP7 exhibits 66%, 69%, and 70% amino acid identity to BUP1, BUP4, and BUP6, respectively, but had no discernable effect on alkaloid levels when evaluated in an engineered yeast context (Fig. 3). Unlike that of BUP1, BUP4, and BUP6, BUP7 gene expression is nearly absent in latex and very low across all other organs (Fig. 6). BUP8 and BUP5 are both highly expressed in the root, while phylogenetic analysis revealed their close clustering (Supplemental Fig. S3). However, the overall expression of BUP5, which shows functionality in yeast, is greater than that of BUP8 (e.g. >2-fold relative transcript abundance in root; Fig. 6). Although BUP2 and BUP9 are expressed in the stem, these genes display a relatively low expression level. BUP3 expression was not detected and could not be amplified from the appropriate cDNA, so it was not included in the analysis. Subcellular localization of BUP1 was determined by bombarding onion epidermal cells with gene expression cassettes, and analysis was carried out by confocal microscopy. This gene was chosen for the localization experiment owing to its prominent clustering profile within the genome, its activity with a broad suite of alkaloids, and its high expression in latex tissue (the site of opiate alkaloid accumulation in opium poppy). BUP1 localized to the plasma membrane (Fig. 7), as confirmed by colocalization with a plasma membrane marker (plasma membrane intrinsic protein [PIP]2A-mCherry).
Figure 6.
Relative expression of BUP1, BUP2, and BUP4–BUP9 across six organs in opium poppy ‘Roxanne’ as measured by RT-qPCR. y Axes use a logarithmic scale where appropriate. Values represent the mean ± sd of three biological replicates. Letters above the bars indicate pairwise differences in the mean values, identified by Tukey-Kramer multiple-comparison tests, with alpha set to 0.05 in all cases.
Figure 7.
BUP1 localizes to the plasma membrane of onion epidermis cells. A and B, Fluorescent images of an onion epidermal cell cobombarded with expression cassettes of pUC19-35S::GFP (A) and pUC19-35S::PIP2A-mCherry (B). C, Merged image of A and B revealing relative localization patterns of GFP (cytosolic) and PIP2A-mCherry (plasma membrane) fluorescent signals. D and E, Fluorescent images of an onion epidermal cell cobombarded with expression cassettes of pUC19-35S::PUP1-GFP (D) and p35S::PIP2A-mCherry (E). F, Merged image of D and E showing colocalization of BUP1-GFP and PIP2A-mCherry (plasma membrane). Scale bar = 50 μm.
Silencing of BUPs Reveals Potential Functional Redundancy
Suppression of BUP transcript levels was pursued by virus-induced gene silencing (VIGS) of opium poppy seedlings. Constructs were designed targeting individual genes (BUP1, BUP2, BUP4, or BUP6, respectively) or multiple genes in different combinations (BUP1, BUP2, and BUP 4; BUP1, BUP4, and BUP6; and BUP1, BUP2, BUP4, and BUP6; Supplemental Fig. S14). Relative transcript abundance of the four BUPs for each silencing construct revealed that the targeted gene(s) in each case were significantly suppressed (Supplemental Fig. S15). BIA profiling of successfully silenced opium poppy plants was conducted by liquid chromatography-mass spectrometry (LC-MS) to monitor for perturbations in the accumulation patterns of major alkaloids (morphine, codeine, thebaine, papaverine, and noscapine; Supplemental Fig. S16). As per our routine VIGS procedure, each silencing experiment was conducted on a population of poppy plants cultivated under defined conditions alongside a population of control plants infiltrated with empty vector. This was done to account for inevitable variations in alkaloid content from experiment to experiment, as the VIGS study was performed over the course of several months and in more than one greenhouse chamber. Overall, very little impact on alkaloid levels was observed, even in cases of concomitant cosilencing of more than one BUP. Nonetheless, some effects were noted; for example, BUP2 silencing leads to a significant reduction in papaverine (P = 0.013). Noscapine levels were reduced by the individual targeting of BUP4 (P = 0.053) and the combinatorial silencing of BUP1, BUP4, and BUP6 (P = 0.028) and BUP1, BUP2, and BUP4 (P = 0.008). Morphine, codeine, and thebaine levels were not significantly affected by any of the silencing constructs.
DISCUSSION
BUPs Transport Opiate Alkaloids
The discovery of BUP1 and its homologs represents a critical step forward in alkaloid biosynthesis, as these transporters operate efficiently on opiates, as well as upstream pathway intermediates and metabolic precursors. Identification of BUP1 and BUP1 pseudogene within two genomic regions containing clusters of BIA biosynthetic genes posed sufficient evidence for further exploration of whether BUP1 and related PUP-type proteins featured activity toward alkaloids. Indeed, evidence presented herein shows that BUPs are effective cellular importers of BIAs, and that different BUP homologs display unique substrate acceptance profiles. Some PUP-type proteins display flexibility in terms of substrate acceptance profiles; for example, PUP1 from Arabidopsis is known to accept adenine (Gillissen et al., 2000), transzeatin (Bürkle et al., 2003), and pyridoxine (Szydlowski et al., 2013) when expressed in yeast, while competition assays suggest binding by, or transport of, several purine derivatives, including kinetin, caffeine, and, to some extent, nicotine (Gillissen et al., 2000; Szydlowski et al., 2013). The only known example of a PUP-like homolog with an established role in plant specialized metabolism (other than the BUPs disclosed herein)—tobacco NUP1—efficiently transports its primary substrate but displays varying degrees of activity with a broader range of nitrogenous metabolites. For example, heterologously expressed NUP1 was shown to import nicotine to yeast in addition to the biosynthetically related alkaloids anatabine, hyoscyamine, and scopolamine (Kato et al., 2015). ABC- and MATE-type transporter families have been implicated in the transport of the alkaloid berberine in Coptis japonica (Shitan et al., 2003, 2013; Takanashi et al., 2017). However, this report identifies transporters capable of accepting opiate alkaloids in opium poppy. Furthermore, this discovery represents an important breakthrough for microbial production of important opiates. Deployment of BUP1 in a synthetic biology context enabled the modular coculturing of yeast engineered with BIA metabolic enzymes and the improvement of titers for the important narcotic analgesics codeine and morphine.
Specialization and Redundancy in the BUP Family
Characterization of opium poppy BUPs in engineered yeast revealed that the transporters accept a wide range of BIA substrates. BUP1 drastically increases the cellular concentrations of yeast fed with dopamine and BIAs, including (S)-reticuline, salutaridine, thebaine, codeine, and morphine (Fig. 4). BUP1 appears to be an effective “generalist” transporter of BIAs, whereas other members display a preference for certain segments of the pathway from dopamine to morphine. For example, BUP2 shows strong uptake activity with the “early” intermediates norcoclaurine, (S)-reticuline, and (R)-reticuline, while BUP6 is very active in the uptake of both norcoclaurine and the morphinan alkaloids (thebaine, codeine, and morphine). Beyond substrate preference profiles, gene expression results similarly reveal both overlapping and tissue-specific expression patterns (Fig. 6). For example, BUP1, BUP4, and BUP6 are highly expressed in latex. In contrast, BUP5 and BUP8 display the highest expression in root tissue. Expression patterns are mirrored in phylogenetic clustering results. For example, BUP1, BUP4, and BUP6 form a monophyletic group, as do BUP5 and BUP8 (Supplemental Fig. S3). This overlap in substrate range and transcript levels, even within a single tissue (e.g. latex) reveals functional redundancy in the BUP family. This redundancy renders in planta knockdown techniques such as VIGS especially challenging tasks, as each BUP sequence is sufficiently different that cosuppression is extremely difficult (Supplemental Fig. S11; Supplemental Dataset S1). For this reason, we opted to design VIGS experiments such that several BUP genes (up to four) could be silenced at once (Supplemental Fig. S14). We targeted latex-expressed genes BUP1, BUP4, and BUP6 for silencing, as latex is the site of opiate accumulation (Onoyovwe et al., 2013). We also included BUP2 in the study, as this gene exhibits a region of high sequence identity enabling convenient cosuppression. While this approach allowed significant reduction in the target transcript (Supplemental Fig. S15), levels of morphinan alkaloid were not significantly impacted (Supplemental Fig. S16). It is possible that even with substantial overall reduction in gene expression, sufficient transcript remained for one or more latex-specific BUPs to prevent an observable diminishment in opiate levels. VIGS-mediated gene suppression only offers a snapshot at a single time point, which might hide the real-time impact of e.g. transporter knockdown and a slower rate of metabolite translocation. Additionally, our analysis focused on major alkaloids within whole-plant extracts rather than examining alkaloid levels in individual tissues, potentially masking tissue-specific perturbances in intermediate alkaloid levels. For example, silencing of stem-predominant BUP2 (Fig. 6) could have impacted stem-specific BIA intermediates of the opiate pathway. Subtle changes in pathway intermediates, which are normally present at low levels in latex-free stem tissues, may have been masked by the overwhelming presence of latex-specific end products. Beyond plant-based VIGS as a functional genomics platform, future work could make use of elicitor-treated opium poppy cell cultures (Zulak et al., 2009) or cultivars with different alkaloid chemotypes (Desgagné-Penix et al., 2012) to investigate potential BUP contribution to alkaloid-specific defense responses and/or variable alkaloid accumulation profiles.
In the metabolite conversion assays, the data suggested specialization of BUP members in the uptake of alkaloid intermediates and end products. Depending on the supplemented metabolite, different BUP-expressing strains fared better. In strain 1, BUP5 and BUP6 outperformed the others when fed l-DOPA or dopamine, while BUP1 improved titers with supplementation of NLDS. The latter result was predictable, as BUP1 is one of the few transporters with efficient cellular uptake of the noncanonical NLDS substrate. Therefore, BUP activity with individual substrates can inform metabolic engineering of the pathway in microbes by guiding either the choice of intermediate supplementation or the choice of BUP isoforms for each module. In strains 2 and 3, BUP1, BUP4, BUP5, and BUP6 were recurring facilitators of improved titers. BUP3 was a poor performer in all but the last metabolite conversion assays; it did confer improved morphine titers to strain 3 fed codeine. This was expected due to its poor substrate acceptance range, especially with dopamine, salutaridine, thebaine, and codeine. However, BUP3 does display a preference for the plant endogenous substrate NCC, which was not included in the conversion assays.
BUP Discovery Opens New Lines of Inquiry for Alkaloid Metabolism
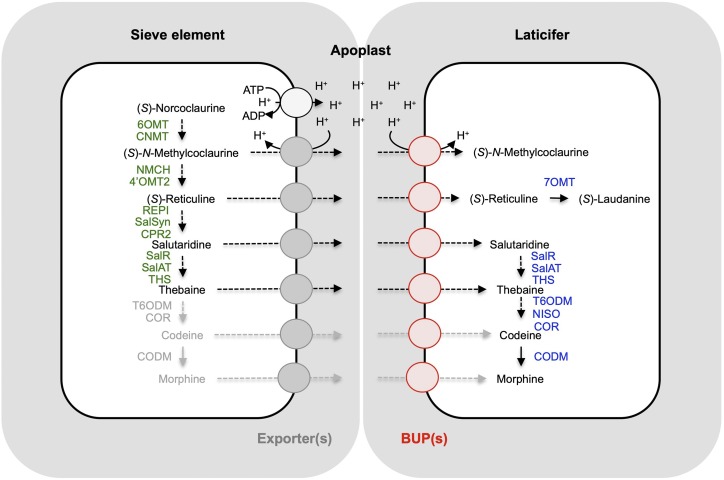
Despite the considerable advancement in our understanding of opiate biosynthesis, the cellular transport of BIAs and their upstream precursors has not been extensively investigated. Indirect evidence has long pointed to the requirement for cellular transport within opium poppy (Bird et al., 2003; Weid et al., 2004; Onoyovwe et al., 2013). Immunofluorescence labeling and shotgun proteomics supported a biosynthetic model in which initial steps of opiate biosynthesis occur in sieve elements and later steps occur in neighboring laticifer cells (Onoyovwe et al., 2013). It was recognized in that previous work that alkaloid translocation from sieve elements to laticifers could occur via symplastic transport through plasmodesmata or apoplastic movement mediated by one or more transporters. The predominance of the BUP1 transcript in latex (Fig. 6), its ability to import a variety of opiate pathway intermediates and link together such pathways in yeast cells (Figs. 4 and 5), its localization to the plasma membrane in plant cells (Fig. 7), and its close association with late-stage biosynthetic enzymes in the opium poppy genome (Supplemental Fig. S1) all point to its likely involvement as a latex-specific importer (Fig. 8). While BUP1 is the only homolog with a sequence traceable to a genomic region encoding laticifer-specific enzymes SalR, SalAT, and THS (Onoyovwe et al., 2013; Chen et al., 2018), the possible involvement of other BUPs in cellular transport must be considered as well. If translocation of intermediates occurs through the apoplast, export from sieve elements would likely be required. An export-import transport system operating across the apoplast was recently characterized in Arabidopsis. Specifically, an ABC transporter exports cytokinins from their biosynthetic site in roots and loads them into the xylem, followed by import through a plasma membrane-localized PUP into the cytosols of aerial organ (or embryonic) cells acting as “cytokinin sinks” (Ko et al., 2014; Zürcher et al., 2016; Kang et al., 2017). A comparable system could conceivably operate in opium poppy, with BUPs acting as importers for “opiate sink” laticifer cells. Interfamily transporter partnerships have been proposed for nicotine and berberine biosynthesis (Shitan et al., 2003, 2013; Morita et al., 2009; Hildreth et al., 2011). The capacity for BUPs in opium poppy to operate not only on final metabolic products (e.g. opiates) but also on pathway intermediates is not unique in plant secondary metabolism. For example, the frequently broad substrate acceptance profiles of NPF and ABC transporter proteins enables their activity with a multitude of metabolites, including pathway intermediates (Corratgé-Faillie and Lacombe 2017; Shitan 2016).
Figure 8.
Proposed role for BUP transporters in opium poppy BIA biosynthesis. Opiate metabolic enzymes are largely restricted to either sieve elements or adjacent laticifers (Onoyovwe et al., 2013). The major alkaloids of the latex include the opiates thebaine, codeine, and morphine, in addition to minor amounts of intermediates (S)-N-methylcoclaurine, (S)-reticuline, and salutaridine. However, initial steps of BIA biosynthesis occur in the sieve elements, implying a transport mechanism to laticifers. Laticifer-localized, proton symport-mediated BUP activity (red) may act in concert with an unknown transport system (gray) to export alkaloid intermediates from the sieve element to the apoplast. Enzymes in green have been localized to sieve elements, whereas enzymes in purple are predominant in latex. Enzymes and alkaloids in gray are not detected in latex-free plant extract (Dastmalchi et al., 2019). 6OMT, norcoclaurine 6-O-methyltransferase; CNMT, coclaurine N-methyltransferase; NMCH, N-methylcoclaurine 3′-hydroxylase; 4′OMT, 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase; CPR, cytochrome P450 reductase; NISO, neopinone isomerase; COR, codeinone reductase.
Beyond opiates, members of the BUP transporter family might be involved in disparate branches of the BIA metabolic network. For example, while BUP7–BUP9 showed marginal or no significant impact in our metabolite conversion assays, root-specific expression of BUP8 suggests uptake activity with alternative metabolites, such as sanguinarine. Morphine and sanguinarine share a common biosynthetic origin with the formation of (S)-reticuline (Fig. 1), but their biosynthetic pathways diverge thereafter. While morphine accumulates in the latex of aerial organs, sanguinarine is a major root alkaloid (Facchini and De Luca, 1995). BUP8 characterization can be expanded to canvass metabolites from (S)-scoulerine to sanguinarine to investigate a putative role in root alkaloid metabolism. Induction of sanguinarine biosynthetic machinery is known to occur in opium poppy cell cultures by application of fungal metabolites (Facchini and Park, 2003), while the benzophenanthradine alkaloid pathway is elicited in cultures of Eschscholzia californica through application of hormones (e.g. jasmonate; Gündlach et al., 1992). Both root and elicited poppy cell cultures could serve as model platforms to investigate putative BUP involvement in inducible defense responses. For example, a study of whether BUP genes are induced in response to environmental cues, as are many other BIA genes (Facchini and Park, 2003), could reveal their role in overall alkaloid metabolism.
The 21-member PUP-type transporter family in Arabidopsis reveals a similar evolutionary trajectory of functional redundancy and divergence (Jelesko, 2012). Two closely related proteins, AtPUP1 and AtPUP2 (64% identity), share substrates (adenine and cytokinins), albeit with different kinetic parameters (Gillissen et al., 2000), while a more distant member, AtPUP3, does not accept purine-derived substrates (Bürkle et al., 2003). Patterns of expression in the Arabidopsis family are also very specialized, e.g. with AtPUP3 restricted to pollen tissue. Alternatively, it is possible that activity on opiates for BUP7–BUP9 was not detected owing to failed expression of these variants. Our attempts to detect yeast-expressed BUPs by immunoblotting failed regardless of whether activity was observed, presenting a caveat in interpretation. While integral membrane proteins can be notoriously difficult to observe using conventional blotting techniques, specialized methods involving GFP fusions and fluorescence detection have been developed for membrane proteins expressed in yeast (Drew and Kim, 2012). Application of these techniques to the study of BUPs, and PUPs generally, could reveal additional functionality for transporters normally displaying recalcitrant expression.
Beyond the nine BUP members discussed in this work, other PUP-type proteins in opium poppy might be involved in the transport of BIAs. For example, noscapine and papaverine, which were not accepted by the BUP homologs examined in this work, may undergo transport via distantly related proteins. Possible candidates include BUP1 homologs with <30% identity, designated transport proteins in the phylogenetic analysis (Supplemental Fig. S3). These proteins share homology with Arabidopsis PUPs, indicating possible roles in the transport of purine alkaloids or derivatives (Zürcher et al., 2016). As discussed above, transport of BIAs might be mediated by members of other protein families (e.g. ABC, MATE, NPF; Morita et al., 2009; Shitan et al., 2014; Payne et al., 2017).
Metabolic Engineering and Modular Polycultures for Efficient BIA Biosynthesis
Inclusion of BUP1 had a dramatic impact when used in the context of cocultured, engineered yeast strains. Remarkably, with the aid of BUP1, the yeast were able to uptake exogenously fed l-DOPA and perform a 14-step bioconversion through over a dozen heterologously expressed catalysts to yield 3 ± 4 mg L−1 OD−1 codeine (Fig. 5C). Opiates were only produced in yeast expressing BUP1, as neither codeine nor thebaine were observed in empty vector control cultures. Furthermore, titers of the promorphinan intermediate salutaridine were elevated ∼10-fold in the presence of BUP1. Demonstration of BUP1’s ability to uptake a broad suite of BIA intermediates (Fig. 4) implies that part of this transporter’s striking impact in a fermentation-based context relies on reuptake of pathway intermediates inadvertently effluxed by endogenous yeast mechanisms. Most BIAs bear a positive charge at pH at or below ∼8, owing to a heterocyclic nitrogen with a relatively high pKa (i.e. the pKa of codeine is 10.6 at 25°C; (Haynes, 2014). Thus, it is expected that pathway intermediates and products would bear a positive charge in yeast media, possibly reducing their membrane permeability. Nitrogenous pharmaceuticals such as sertraline are known to be internalized across S. cerevisiae membranes through passive diffusion, but only when bearing a neutral charge (Chen et al., 2012). Regardless of whether uptake is passive or transporter aided, the process is likely counteracted by energy-dependent, endogenous, and broad-specificity xenobiotic efflux pumps (e.g. ABC transporters; Thakur et al., 2008). Thus, without an alkaloid-specific uptake permease, pathway intermediates are likely lost to the surrounding media without the ability to re-enter the cell and finish bioconversion. We chose to include BUP1 in this coculturing exercise owing to its ability to import a broad suite of alkaloids along the opiate pathway (Fig. 4). However, results showing some degree of “specialization” among the BUP variants toward certain alkaloids could justify the use of specific BUPs for specific engineering purposes. For example, accumulation of intermediates in favor of opiate end products could potentially be aided by using alternative BUP variants favoring the import of early-stage over later-stage pathway metabolites (e.g. BUP2 or BUP3; Fig. 4).
Metabolic engineering of heterologous pathways into fermentation-based platforms generally involves the incorporation of genes from one or more source organisms into a single microbial strain to enable complete bioconversion of substrate to product. Monoculture fermentation is challenged by the requirements of integrating increasingly complicated pathways within a single cellular context. A growing number of studies have leveraged the idea of a modular coculture engineering strategy (Zhang and Wang, 2016). A biosynthetic pathway is divided into separate, serial modules, each incorporated into individual microbial strains, grown together in a single fermentation. This strategy has been employed successfully in the microbial biosynthesis of plant-derived compounds including taxanes (Zhou et al., 2015), monolignols (Chen et al., 2017), and flavonoids (Jones et al., 2016). Our modular coculturing strategy with opiate production in yeast suggests that BUP1 facilitates the shuttling of intermediates between tandem cultures (Fig. 5). Inclusion of the transporter in multistrain cocultures improved titers of salutaridine and thebaine, and it enabled the biosynthesis of codeine.
Modular coculturing reduces metabolic burden on the host microbe in overexpression of heterologous proteins or production of de novo chemicals. Coculturing distributes the labor among multiple strains and allows the use of microbes of different genetic or cellular background. An E. coli-S. cerevisiae coculture has been used for taxol precursor biosynthesis and production of BIAs (magnoflorine and corytuberine; Minami et al., 2008; Ajikumar et al., 2010). Segmentation also improves module insulation from undesired genetic, regulatory, or inhibitory interactions, or promiscuous enzyme activity (Chen et al., 2017). In the case of our polycultured strains (Fig. 5), separation of the pathway into three modules may reduce negative feedback of metabolites (e.g. (S)-reticuline or thebaine) on untargeted enzymes. For example, Tyr hydroxylase has been shown to be inhibited by both substrate (Tyr) and product (catecholamine; Trenchard et al., 2015) in yeast; NCS has been characterized with product (NLDS) inhibition (Samanani et al., 2004). Perhaps rapid efflux of NLDS by a novel/endogenous transporter, followed by BUP1 uptake into a separate module could relieve negative inhibition in strain 1 and unlock further efficiencies.
Bioconversion of l-DOPA to thebaine and codeine in the multistrain cocultures revealed a bottleneck after reticuline production (Fig. 5). The inclusion of BUP1 improved titers of the target products but did not promote significant consumption of the initial pool of reticuline. Catalytic inefficiencies appear to exist in the turnover of (S)-reticuline (by REPI) and (R)-reticuline (by SalSyn). These bottlenecks were apparent in the single-module conversions (e.g. strain 2; Fig. 3), where (S)-reticuline supplementation was swapped for (R)-reticuline and then salutaridine, recording drastic incremental improvements with each bypassing of an inefficient enzyme. One way to deal with this problem, in lieu of finding alternative catalysts, is to alter the strain-strain ratios (Jones and Koffas, 2016). Favoring strain 2 (or strains 2 and 3) could improve consumption of (S)-reticuline produced by strain 1. Modular coculturing enables plug-and-play biosynthesis of various target products or high-throughput screening of strain variants. For example, our platform can be modified by swapping strain 3 for a new module capable of transforming thebaine to various N-demethylated derivatives. BUP1 inclusion in the new module would provide strong thebaine uptake capacity. Finally, it is worth considering that although BUP1 is plasma membrane localized in planta (Fig. 7), such localization may not necessarily occur in yeast. Intracellular localization of BUPs and other heterologously expressed pathway components could possibly serve to illuminate unforeseen, transport-based pathway bottlenecks and provide a basis for engineering improvements.
CONCLUSION
BUPs represent a key element within BIA metabolism, establish a tangible lead in our understanding of how the poppy plant expands its biosynthetic pathway across at least two cell types, and allow further probing into the evolution of the PUP-derived transporter family. The cellular uptake activity of BUPs, with substrates ranging from dopamine to morphine, expands the role of PUP-type transporters in specialized metabolism beyond purine-derived compounds. Several BUPs improve titers of intermediates and end-products in multistep conversion of l-DOPA, (S)-reticuline, and thebaine, presumably by increasing cellular concentrations. When the opiate biosynthetic pathway enzymes were stitched together by modular coculturing and fed l-DOPA, the inclusion of BUP1 improved titers of salutaridine and thebaine and led to the formation of codeine. Deployment of BUPs can register further improvements of titers, rate, and yield, building on the achievements recorded with the addition of new opiate enzymes, THS and neopinone isomerase (Chen et al., 2018; Dastmalchi et al., 2019).
MATERIALS AND METHODS
Chemicals and Plant Cultivation
(S)-Reticuline was a gift from Tasmanian Alkaloids. Morphine and codeine were gifts from Sanofi. Salutaridine and thebaine were purchased from Kalexsyn and Toronto Research Chemicals, respectively. Noscapine and papaverine were purchased from Sigma-Aldrich. Other chemicals and reagents were purchased from Sigma-Aldrich, unless otherwise noted. Neopine and neomorphine were produced by enzymatic conversion as previously described (Dastmalchi et al., 2019). Opium poppy (Papaver somniferum) plants were cultivated in growth chambers at 20°C/18°C (light/dark) with a photoperiod of 16 h provided by a combination of cool-white fluorescent and incandescent lighting. All research involving controlled materials was performed with appropriate government approval.
BUP Identification and Linkage Mapping of Genomic Data
BUP1, which was previously identified as a pseudogene on the genomic scaffold containing SalR, SalAT, and THS (Chen et al., 2018), was used to query (1) extensive opium poppy transcriptomic resources (Desgagné-Penix et al., 2010; Chen et al., 2018) and (2) a recently assembled draft genome of opium poppy (Dastmalchi et al., 2019). Transcriptomic sequences with >50% sequence identity to BUP1 were compiled to arrive at a list of five full-length clones (BUP1–BUP6). An additional selection of BUP transcripts with complete coding sequences exhibiting between 30% and 50% sequence identity to BUP1 were identified as BUP7–BUP9. cDNA and amino acid sequences for BUP1–BUP9 are found in Supplemental Datasets S1 and S2, respectively. BUP1 through BUP9 were used as queries for genome BLASTing to locate genomic scaffolds with cognate genes (100% matches), “BUP-like” genes, or partial/pseudo BUP genes. These genomic scaffolds were further subjected to gene, coding sequence, and ORF prediction analysis using the Augustus feature of Geneious v. 11.1.2, with Arabidopsis (Arabidopsis thaliana) tool settings (Stanke et al., 2004). The putative coding sequences of linked genes were BLASTed against publicly available National Center for Biotechnology Information databases to assign functional annotations.
Phylogenetic Analysis
Protein sequences of BUP1–BUP9 (Supplemental Dataset S2) were aligned with functionally characterized PUP- or NUP-type transporters. Additional BUP homologs from opium poppy transcriptomes exhibiting low sequence identity with BUP1 (<30%) were included in the analysis. Abbreviations and GenBank accession numbers for previously characterized permease transporters are as follows: Arabidopsis AtPUP1 (Q9FZ96), AtPUP2 (Q94GB1), AtPUP3 (Q9FZ95), AtPUP7 (Q2V3H2), AtPUP14 (Q9FXH5), and Nicotiana tabacum NtNUP1 (ADP30798). Oryza sativa homologs OsPUP4 and OsPUP7 are found as loci LOC_Os01g48800 and LOC_Os05g48300 in the National Science Foundation Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). Alignment of protein sequences was performed using default MUSCLE tool settings of EMBL-EBI (https://www.ebi.ac.uk) with Phylip Interleaved output format. Alignments formed the basis for tree generation using PhyML 3.0 (Guindon et al., 2010) and MrBayes 3.2.6 (Ronquist et al., 2012) for trees shown in Supplemental Figure S3. Akaike Information Criterion was used with the maximum likelihood-based method aLRT (approximate likelihood ratio test, SH-like algorithm) and 100 iterations for bootstrap calculations. Numbers at nodes represent bootstrap values. Tree visualization was performed using iTOL v.3 (Letunic and Bork, 2007).
Yeast Uptake Assays
Yeast (Saccharomyces cerevisiae) strain CEN.PK102-5B was transformed with a Gal-inducible, transient expression construct based on pESC-His. This plasmid had been modified with the addition of a gene conferring antibiotic resistance to G418 (KanMX; Walker et al., 2003) as described (Dastmalchi et al., 2019). Codon-optimized coding sequences for BUP1–BUP9 including C-terminal c-myc epitope tags were synthesized at GenScript USA (www.genscript.com) and subcloned to multiple-cloning site 1 of the modified pESC-His using BamHI and SacII restriction sites. Following transformation, four individual colonies were selected per clone for yeast uptake assays. Immunoblotting was performed as described (Dastmalchi et al., 2019) for detection of BUP-fused c-myc epitope, although no signal was observed regardless of the BUP variant examined. Uptake assays were conducted as follows: yeast strains transiently expressing one of BUP1–BUP9 or no gene (“empty vector” controls) were inoculated in 200 μl synthetic dropout (SD) medium containing 2% (w/v) Glc overnight in a 96-well format, using a Fisher brand Incubating Microplate Shaker (Fisher Scientific). The overnight cultures were then diluted with 300 μl SD medium containing 2% (w/v) Gal and 1 mm l-DOPA or dopamine, or 200 μm of alkaloid substrate. Yeast cultures were grown for 24 h postinduction at 30°C unless otherwise noted. An exception is the case of the time-course uptake experiment, where yeast underwent incubation for 12, 24, 48, or 72 h. Following culturing, yeast cells were gently pelleted and washed twice with media. l-DOPA, dopamine, and BIAs in yeast cell pellets were extracted twice with 500 μL of 100% (v/v) methanol. Insoluble matter was centrifuged, and the supernatant was pooled, dried, and resuspended in 200 μL of 50:50 water:acetonitrile (ACN). Five µL aliquots were subjected to high-resolution MS analysis.
Construction and Fermentation of Yeast Strains for Metabolic Conversion Assays
The parent yeast (S. cerevisiae) strain for engineering through genomic integration was CEN.PK102-5B. Integration cassettes were assembled based on a series of plasmids first reported by Mikkelsen et al. (2012) and improved upon by Jensen et al. (2014). These plasmids were generously provided as a gift by Dr. Irina Borodina (Technical University of Denmark), Dr. Uffe Hasbro Mortensen (Technical University of Denmark), and Dr. Barbara Ann Halkier (University of Copenhagen). Construct assembly, genomic integration, and fermentation were performed as described previously (Morris et al., 2016). Briefly, coding sequences for biosynthetic genes were codon-optimized and synthesized at GenScript USA (www.genscript.com), followed by subcloning to integration vectors using standard, USER-based strategy (Supplemental Table S1; Salomonsen et al., 2014). Each integration vector hosted two biosynthetic genes under control of a bidirectional, inducible promoter system (Gal1/Gal10). Three plasmids were used to engineer strain 1: (1) Pseudomonas putida DODC plus Homo sapiens MAO-A; (2) opium poppy NCS plus opium poppy norcoclaurine 6-O-methyltransferase; and (3) opium poppy coclaurine N-methyltransferase plus opium poppy 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase. Three plasmids were used to engineer strain 2: (1) opium poppy REPI plus N-terminally truncated (∆9) opium poppy SalSyn; (2) opium poppy SalR plus opium poppy SalAT; and (3) opium poppy THS plus opium poppy cytochrome P450 reductase 2. Two plasmids were used to engineer strain 3: (1) opium poppy T6ODM plus opium poppy codeinone reductase 1.3; and (2) opium poppy codeine O-demethylase. Stepwise genomic integration of these plasmids to strains 1–3 was done as previously described (Morris et al., 2016). Each of these engineered strains was then transformed with a transient expression construct with no gene (empty vector control) or one of BUP1–BUP9. The transient expression construct was a modified version of the Gal-inducible plasmid pESC-His, exhibiting an additional selection marker (KanMX) for antibiotic G418 resistance (Dastmalchi et al., 2019). Plasmids hosting BUPs or no gene (controls) were individually transformed to the platform yeast strains with chromosome-integrated BIA biosynthetic genes using the LiAc/polyethylene glycol/single-stranded carrier DNA transformation method, and four resulting transformants were selected for each clone as biological replicates. Yeast strains transiently expressing candidate BUP genes were inoculated in 200 μL SD medium containing 2% (w/v) Glc overnight, in a 96-well format, using an incubating microplate shaker (Fisher Scientific). The overnight cultures were then diluted with 300 μL SD medium containing 2% (w/v) Gal and 250 μm alkaloid substrate for bioconversion. For l-DOPA and dopamine feeding, concentrations of 1 mm were used in the media. Yeast cultures were grown for 24 h postinduction at 30°C. Yeast cells were removed by centrifugation, and 5 μL of supernatant, containing metabolites secreted by the yeast cells into the culture medium, was subjected to high-resolution MS analysis. For our experiment involving strain 1-based bioconversion of exogenously fed NLDS and follow-up analysis of both media and cellular lysates (Supplemental Fig. S6), the same procedures were employed, with the exception that postincubation, the yeast cells removed by centrifugation were retained for alkaloid analysis. Lysis, extraction, and analysis were performed as described for yeast uptake experiments.
Cofermentation of Yeast Strains 1–3 Transiently Expressing BUP1
S. cerevisiae strains 1 and 3, with chromosome-integrated BIA biosynthetic genes and transient expression of BUP1 in modified pESC-His, were individually inoculated in 200 μL SD medium containing 200 μg/L of G418 and 2% (w/v) Glc and cultured overnight at 30°C. The same was done for strain 2, except that this strain was transiently expressing modified pESC-His harboring both BUP1 (in MCS2) and opium poppy THS (in MCS1; Chen et al., 2018) to support sufficient thebaine formation. The overnight cultures of strains 1–3 were then cocultured into 300 μL of SD medium containing 2% (w/v) Gal and 200 μg/L of G418 and 1 mm of l-DOPA to allow bioconversion. Yeast cocultures were grown for 24 h postinduction at 30°C, followed by removal of cells by centrifugation. Five μL of the media supernatant, containing metabolites secreted by the yeast cells into the culture medium, was subjected to high-resolution MS analysis.
High-Resolution MS for In Vivo Yeast Assays
Yeast media and cell extracts were analyzed by high-resolution LC-electrospray ionization (ESI)-linear trap quadrupole (LTQ)-Orbitrap-XL MS (Thermo Fisher Scientific). Cell extracts were preprocessed prior to loading to remove protein by 50:50 dilution with methanol, centrifugation to pellet insolubles, and retrieval of supernatant for analysis. Samples were either loaded directly or diluted with methanol to ensure target metabolites remained within linear ranges as indicated by standard curves using authentic standards. High-resolution LC-ESI-LTQ-Orbitrap-XL MS (Thermo Fisher Scientific) was performed using a modified version of a method described previously (Chang et al., 2015), with the exception that liquid chromatography was carried out using an UltiMate 3000 HPLC (Thermo Fisher Scientific) equipped with a Poroshell 120 SB-C18 column (Agilent Technologies) instead of an Accela HPLC system (Thermo Fisher Scientific) equipped with a Zorbax C18 column (Agilent Technologies). Five μL of samples were fractionated at a flow rate of 0.5 mL/min and a gradient of solvent A (10 mm ammonium acetate, pH 5.5, 5% [v/v] ACN) and solvent B (100% [v/v] ACN) as follows: 100% to 80% (v/v) solvent A over 5 min; 80% to 50% (v/v) over 3 min; 50% to 0% (v/v) over 3 min; isocratic at 0% (v/v) for 2 min; 0% to 100% (v/v) over 0.1 min; and isocratic at 100% (v/v) for 1.9 min. Total run time was 15 min, but data were collected for only 10 min. Heated ESI source and interface conditions were operated in positive ion mode as follows: vaporizer temperature, 400°C; source voltage, 3 kV; sheath gas, 60 au, auxiliary gas, 20 au; capillary temperature, 380°C; capillary voltage, 6 V; tube lens, 45 V. LTQ-Orbitrap-XL (Thermo Fisher Scientific) instrumentation was performed as three scan events in data-dependent, parallel-detection mode. The first scan consisted of high-resolution Fourier transform MS from 150 to 450 m/z with ion injection time of 500 ms and scan time of ∼1.5 s. The second and third scans (∼0.5 s each) collected collision-induced dissociation (CID) spectra in the ion trap, where the parent ions represented the first- and second-most abundant alkaloid masses, respectively, as determined by fast Fourier transform preview using a parent ion mass list corresponding to exact masses of known alkaloids. Dynamic-exclusion and reject-ion-mass-list features were enabled. External and internal calibration procedures ensured <2 ppm error. Exact mass, retention times, and CID spectra of authentic standards were used to identify alkaloids, and quantification was performed using standard curves. The Quan Browser feature of XCalibur v. 3.1 (Thermo Fisher Scientific) was employed for automated peak identification and quantification.
RNA Extraction, cDNA Synthesis, and RT-qPCR
Total RNA from frozen opium poppy tissue samples (‘Roxanne’), finely ground using a TissueLyser (Qiagen), was extracted using cetyl trimethyl ammonium bromide. Latex samples were harvested from opium poppy flower buds 1–2 d before anthesis. The stem was cut ∼1 cm below the bud and the exuding latex was collected. Stem samples were harvested by collecting a 2- to 3-cm length of stem below the bud, dissected into thin slices, and placed in water to drain latex. cDNA synthesis was performed in a 10-μL reaction containing ∼1 μg of total RNA using All-in-One RT master mix (ABM) according to the manufacturer's instructions. Amplification was performed using SYBR Green master mix (Applied Biosystems) and was verified by melt curve analysis on a QuantiStudio Real-Time PCR System 3 (Applied Biosystems). Gene-specific primers were used for all RT-qPCR experiments (Supplemental Table S2). All samples were analyzed in technical triplicates. The 2-ΔΔCt method was used to calculate the relative transcript abundance, whereby the mean of the polyubiquitin-10 and actin reference genes served to normalize BUP expression. PCR amplification efficiency, calculated from raw fluorescence data using LinRegPCR (Ramakers et al., 2003) was between 90% and 100% for all primer pairs.
Construct Assembly for BUP1 Subcellular Localization
BUP1 cDNA was inserted between a cauliflower mosaic virus 35S promoter and Aequoria victoria GFP of the pUC19-35S::GFP vector via the sequence- and ligation-independent cloning method (Jeong et al., 2012). All primers used in construct assembly are listed in Supplemental Table S2. In brief, pUC19-35S::GFP was linearized via reverse PCR using a set of primers annealing to the end of the 35S promoter (reverse direction) and the beginning of GFP (forward direction). The BUP1 cDNA was amplified by PCR using a set of primers with 5′ extensions of 20 bp homologous to each end of the linearized vector. The linearized vector and the BUP1 insert were mixed at a molar ratio of 1:3 with T4 DNA polymerase at room temperature for 2.5 min. The mixture was put on ice for 10 min and then transformed to Escherichia coli TOP10 competent cells (Thermo Fisher Scientific). Plant organelle fluorescent marker plasmid pm-rk CD3-1007 (plasma membrane protein PIP2A-mCherry in binary vector pBIN20 with the 35S promoter; Nelson et al., 2007) was ordered through the Arabidopsis Biological Resource Center (https://abrc.osu.edu). Due to the low copy number of pBIN20 in E. coli and the high quantity of plasmid required for the subsequent bombardment experiment, the PIP2A-mCherry cassette was reamplified and ligated into the pUC19 vector using the sequence- and ligation-independent cloning method as described above.
Microprojectile Bombardment of Onion Cells
Gold particles (30 mg, 1.6 μm diameter; Bio-Rad) were sterilized by vortexing in 1 mL of 100% (v/v) ethanol for 5 min, washed twice with sterile, distilled water, and resuspended in 0.5 mL of sterile, distilled water. In a 50 μL aliquot of the suspension, 20 μL of 0.1 m spermidine and 15 μg of plasmid DNA (50 μL) were successively added. After each addition, the mixture was sonicated and vortexed, each time for 10 s. Thereafter, 50 μL of 2.5 m CaCl2 was added to the mixture drop by drop, waiting 5 s after each drop, followed by brief vortexing or sonicating . The final mixture was put on ice for 5 min. The gold particles were then washed twice with ethanol with sonicating and resuspended in 45 μL of 100% (v/v) ethanol. For each bombardment, 15 μL of the particle suspension (1 mg of particles per shot) was pipetted onto the center of a microcarrier that was sterilized with 100% (v/v) ethanol and used after all ethanol had evaporated. Onion bulb scale leaves were cut into segments of ∼5 cm × 3 cm. A single piece of cut scale leaf was placed in the center of a 9 cm petri dish with the abaxial surface facing upwards and positioned below a microprojectile-stopping screen. Bombardments were performed using a biolistic particle acceleration device (PDS 1000/He, Bio-Rad) under a chamber pressure of 26 mm Hg, at 1.5, 2.0, and 6.5 cm from the rupture disc to the microcarriers to the stopping screen to the target, respectively, and at a helium (He) pressure of 900 psi. After bombardment, the plate was sealed and stored at room temperature. After 36 h, the epidermal layer was removed and examined for fluorescent signal. Viable cells were observed without fixation. Fluorescent images were taken using a Leica SP5 laser confocal microscope, with an oil immersion 40× objective. An argon light source is used for GFP and a HeNe543 light source is used for RFP, with excitation wavelengths of 488 nm and 543 nm, respectively. The emission range for GFP is 493–538 nm and the emission range for RFP is 585–649 nm; both signals were detected with HyD detectors. Viable cells were observed without fixation. Fluorescent images were captured using a Leica DM RA2 microscope (Leica Microsystems) with an A4 filter, a Retiga EX digital camera (QImaging), and Open Lab software (version 2.09, Improvision). Microscopic images were captured using the Leica microscope and Retiga camera mounted with a red-green-blue color liquid crystal filter (QImaging).
VIGS
Gene silencing was performed using the tobacco rattle virus (TRV) vector system, which is based on the pYL156 plasmid, as described previously (Hagel and Facchini, 2010). Seven pTRV-RNA2 (pTRV2)-based constructs were designed to specifically silence BUP1, BUP2, BUP4, and BUP6, while an additional three constructs were designed to cosilence the expression of multiple BUP genes. Regions of ∼300–600 bp representing one or more segments of targeted genes (Supplemental Fig. S14; Supplemental Table S3) were synthesized at GenScript and subcloned into pTRV2 using XbaI and Kpn1. Agrobacterium tumefaciens strains harboring (1) pTRV1 and (2) one of seven pTRV2-based silencing constructs or the empty pTRV2 vector were coinoculated at a 1:1 ratio into the cotyledons and apical meristem of 2-week-old opium poppy, Bea’s choice variety, using a needleless syringe. Infiltrated plants were harvested after 8–12 weeks of growth and stored at −80°C. Each plant was then ground to a fine powder using liquid N2 and the aid of a TissueLyser II (Qiagen) instrument at 30 Hz for 1 min with a precooled 30 mL grinding jar and a 10‐mm stainless steel ball. For each plant, the resulting fine powder was divided into two analysis streams consisting of either (1) cDNA synthesis for PCR and RT-qPCR or (2) extraction for metabolite analysis. For the former cDNA stream, powder was extracted for RNA (Dastmalchi et al., 2019) followed by cDNA synthesis. Samples were screened by PCR to identify those derived from successfully infected plants, as evidenced by PCR product specific to the sequence of pYL156 plasmid (GAPDH; Supplemental Table S2). For each VIGS construct, ∼10–12 samples (derived from 10–12 plants) were selected based on positive PCR results. The cDNA was then analyzed using RT-qPCR to determine relative BUP transcript abundance using gene-specific primers (Supplemental Table S2). For the metabolite analysis stream, frozen powder was suspended in a 1 mL solution of methanol and ACN (1:1) in a preweighed 1.5 mL tube. Samples were incubated on a shaker at room temperature and 200 rpm for 4 h. The insoluble matter was removed by centrifugation, and the supernatant was removed to a fresh tube. The insoluble matter was dried and weighed, allowing the 1-mL supernatants to be adjusted to a final concentration of 25 μg (dry weight)/μL.
LC-MS for VIGS Experiments
VIGS sample analysis was conducted using an Agilent 1200 HPLC coupled to an Agilent 6410B triple-quadrupole MS. To remain within linear ranges for each analyte, dilution (1:10) was carried out prior to analysis to measure the more abundant alkaloids (morphine, codeine, thebaine, noscapine, and papaverine). Five-μL aliquots were injected onto a Poroshell 120 SB-C18 HPLC threaded column (Agilent Technologies), with a flow rate of 0.6 mL/min and a gradient of solvent A (10 mm ammonium acetate, pH 5.5, 5% [v/v] ACN) and solvent B (100% [v/v] ACN) as follows: 0% to 60% (v/v) solvent B from 0 to 8 min; 60% to 99% (v/v) solvent B from 8 to 10 min; isocratic 99% (v/v) solvent B from 10 to 11 min; 99% to 0% (v/v) solvent B from 11 to 11.1 min; 0% (v/v) solvent B from 11.1 to 14.1 min. ESI and full-scan mass analyses (m/z range 50–200) and collisional MS/MS multiple-reaction monitoring experiments were performed as described previously (Desgagné-Penix et al., 2012). For high-abundance alkaloids, extracted ion chromatographs of full scans were used. For all other alkaloids, multiple reaction monitoring experiments were conducted using established transitions (Desgagné-Penix et al., 2012). Metabolites were identified and quantified based on comparison of retention times, peak areas, and CID spectra with those of authentic standards at known concentrations. Standard curves were prepared for all alkaloids in the range 0.5–10 μm.
Statistical Analyses
For analyses of plug-and-play metabolic conversion assays, means were compared to those of appropriate metabolite controls (empty vector) using an unpaired, two-tailed Student’s t test allowing P values to be calculated in each case (Fig. 3). Similarly, for analyses of yeast uptake assays and modular coculture data, means were compared to those of appropriate control values (empty vector cultures) using unpaired, two-tailed Student’s t test, with all P values indicated (Figs. 4 and 5, B and C). For analyses of VIGs results, data with multiple series variance were determined using a one-way ANOVA (P < 0.05). Tukey-Kramer multiple-comparison tests were performed to reveal pairwise differences between the means, with alpha set to 0.05 in all cases. For analysis of gene expression and metabolite profiling, statistical significance was determined using a Student’s t test (two-tailed, two-sample, assuming unequal variance).
Accession Numbers
Partially annotated genomic scaffolds harboring BUP genes have been submitted to the National Center for Biotechnology Information under the following accession numbers: MH837996 (scf7180001498639); MH837997 (scf7180001499132); MH837998 (scf7180001499910); MH837999 (scf7180001502001); MH838000 (scf7180001502321); MH838001 (scf7180001503556); MH838002 (scf718000150442); MH838003 (scf718000150530); MH838004 (scf718000150588); MH838005 (scf7180001506323); and MH838006 (scf7180001507141). BUP cDNA sequences are submitted to GenBank with the following accession numbers: MH830312 (BUP1); MH830313 (BUP2); MH830314 (BUP3); MH830315 (BUP4); MH830316 (BUP5); MH830317 (BUP6); MH830318 (BUP7); MH830319 (BUP8); and MH830320 (BUP9).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Opium poppy genomic scaffold scf7180001505888 containing BUP1 pseudogene.
Supplemental Figure S2. Opium poppy genomic scaffold scf7180001505306 containing BUP1.
Supplemental Figure S3. Phylogenetic tree showing relationships between opium poppy BUPs, characterized PUPs, and a characterized nicotine uptake permease.
Supplemental Figure S4. Alignment of opium poppy BUPs and characterized homologous protein sequences.
Supplemental Figure S5. Design for in vivo BUP assays using native or engineered strains of S. cerevisiae.
Supplemental Figure S6. In vivo BUP assays using engineered S. cerevisiae fed NLDS in media and measuring product in both media and cellular lysates postincubation.
Supplemental Figure S7. Opium poppy genomic scaffold scf7180001498639 containing BUP2.1.
Supplemental Figure S8. Opium poppy genomic scaffold scf7180001502001 containing BUP2.2, BUP3.1, and other BUP-like genes.
Supplemental Figure S9. Opium poppy genomic scaffold scf7180001507141 containing BUP3.2 and BUP9.2.
Supplemental Figure S10. Opium poppy genomic scaffold scf7180001506323 containing BUP4.
Supplemental Figure S11. Opium poppy genomic scaffold scf7180001502321 containing BUP5.1 and BUP5.2.
Supplemental Figure S12. Opium poppy genomic scaffold scf7180001499132 containing BUP6.
Supplemental Figure S13. Time course assay for S. cerevisiae expressing BUP1, measuring time-dependent uptake of DOPA, dopamine, and alkaloids.
Supplemental Figure S14. Region(s) of BUP1, BUP2, BUP4, and BUP6 used to prepare VIGS constructs.
Supplemental Figure S15. Suppression of BUP transcript levels in opium poppy plants subjected to VIGS using various constructs.
Supplemental Figure S16. Alkaloid levels in opium poppy plants used in VIGS experiments.
Supplemental Table S1. Primers used to amplify genes and bidirectional Gal1/Gal10 promoter for USER-based cloning into S. cerevisiae genomic integration vectors.
Supplemental Table S2. Primers used for cloning, RT-qPCR analysis, and PCR analysis.
Supplemental Table S3. BUP-derived sequences cloned into pTRV2 for VIGS experiments.
Supplemental Dataset S1. cDNA sequences of BUP genes examined in this study.
Supplemental Dataset S2. Amino acid sequences of BUP proteins used in this study.
Supplemental Dataset S3. Annotation of predicted genes occurring in genomic scaffolds encoding BUPs.
ACKNOWLEDGMENTS
Patent applications related to this work have been filed (PCT/CA2017/050779 and PCT/CA2018/051604).
Received May 13, 2019; accepted August 19, 2019; published August 29, 2019.
Footnotes
This work was supported by funds awarded through the Industrial Research Assistance Program (IRAP; Project 86155) operated by the National Research Council of Canada to Epimeron Inc. Epimeron Inc. is a wholly-owned subsidiary of Willow Biosciences Inc.
References
- Ajikumar PK, Xiao W-H, Tyo KEJ, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330: 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DA, Franceschi VR, Facchini PJ (2003) A tale of three cell types: Alkaloid biosynthesis is localized to sieve elements in opium poppy. Plant Cell 15: 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürkle L, Cedzich A, Döpke C, Stransky H, Okumoto S, Gillissen B, Kühn C, Frommer WB (2003) Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J 34: 13–26 [DOI] [PubMed] [Google Scholar]
- Chang L, Hagel JM, Facchini PJ (2015) Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol 169: 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Korostyshevsky D, Lee S, Perlstein EO (2012) Accumulation of an antidepressant in vesiculogenic membranes of yeast cells triggers autophagy. PLoS One 7: e34024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Hagel JM, Chang L, Tucker JE, Shiigi SA, Yelpaala Y, Chen H-Y, Estrada R, Colbeck J, Enquist-Newman M, et al. (2018) A pathogenesis-related 10 protein catalyzes the final step in thebaine biosynthesis. Nat Chem Biol 14: 738–743 [DOI] [PubMed] [Google Scholar]
- Chen Z, Sun X, Li Y, Yan Y, Yuan Q (2017) Metabolic engineering of Escherichia coli for microbial synthesis of monolignols. Metab Eng 39: 102–109 [DOI] [PubMed] [Google Scholar]
- Corratgé-Faillie C, Lacombe B (2017) Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J Exp Bot 68: 3107–3113 [DOI] [PubMed] [Google Scholar]
- Dang TT, Chen X, Facchini PJ (2015) Acetylation serves as a protective group in noscapine biosynthesis in opium poppy. Nat Chem Biol 11: 104–106 [DOI] [PubMed] [Google Scholar]
- Dastmalchi M, Chen X, Hagel JM, Chang L, Chen R, Ramasamy S, Yeaman S, Facchini PJ (2019) Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat Chem Biol 15: 384–390 [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I, Farrow SC, Cram D, Nowak J, Facchini PJ (2012) Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol Biol 79: 295–313 [DOI] [PubMed] [Google Scholar]
- Desgagné-Penix I, Khan MF, Schriemer DC, Cram D, Nowak J, Facchini PJ (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol 10: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D, Kim H (2012) Screening for high-yielding Saccharomyces cerevisiae clones: Using a green fluorescent protein fusion strategy in the production of membrane proteins. Methods Mol Biol 866: 75–86 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Chen X, Colbeck JC, Tucker J, inventors. (April 1, 2018. Compositions and methods for making benzylisoquinoline alkaloids, morphinan alkaloids, thebaine, and derivatives thereof. United States Patent Application No. PCT/US2017/039589.
- Facchini PJ, De Luca V (1995) Phloem-specific expression of tyrosine/dopa decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7: 1811–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchini PJ, Park S-U (2003) Developmental and inducible accumulation of gene transcripts involved in alkaloid biosynthesis in opium poppy. Phytochemistry 64: 177–186 [DOI] [PubMed] [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen B, Bürkle L, André B, Kühn C, Rentsch D, Brandl B, Frommer WB (2000) A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Gündlach H, Müller MJ, Kutchan TM, Zenk MH (1992) Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci USA 89: 2389–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel JM, Facchini PJ (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat Chem Biol 6: 273–275 [DOI] [PubMed] [Google Scholar]
- Hagel JM, Morris JS, Lee EJ, Desgagné-Penix I, Bross CD, Chang L, Chen X, Farrow SC, Zhang Y, Soh J, et al. (2015) Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol 15: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes WM, editor. (2014) CRC Handbook of Chemistry and Physics. Ed 95. CRC Press, Boca Raton, FL, pp. 5–103 [Google Scholar]
- Hildreth SB, Gehman EA, Yang H, Lu R-H, Ritesh KC, Harich KC, Yu S, Lin J, Sandoe JL, Okumoto S, et al. (2011) Tobacco nicotine uptake permease (NUP1) affects alkaloid metabolism. Proc Natl Acad Sci USA 108: 18179–18184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelesko JG. (2012) An expanding role for purine uptake permease-like transporters in plant secondary metabolism. Front Plant Sci 3: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen NB, Strucko T, Kildegaard KR, David F, Maury J, Mortensen UH, Forster J, Nielsen J, Borodina I (2014) EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res 14: 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J-Y, Yim H-S, Ryu J-Y, Lee HS, Lee J-H, Seen D-S, Kang SG (2012) One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl Environ Microbiol 78: 5440–5443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JA, Koffas MAG (2016) Optimizing metabolic pathways for the improved production of natural products. Methods Enzymol 575: 179–193 [DOI] [PubMed] [Google Scholar]
- Jones JA, Vernacchio VR, Sinkoe AL, Collins SM, Ibrahim MHA, Lachance DM, Hahn J, Koffas MAG (2016) Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng 35: 55–63 [DOI] [PubMed] [Google Scholar]
- Kang J, Lee Y, Sakakibara H, Martinoia E (2017) Cytokinin transporters: GO and STOP in signaling. Trends Plant Sci 22: 455–461 [DOI] [PubMed] [Google Scholar]
- Kato K, Shitan N, Shoji T, Hashimoto T (2015) Tobacco NUP1 transports both tobacco alkaloids and vitamin B6. Phytochemistry 113: 33–40 [DOI] [PubMed] [Google Scholar]
- Ko D, Kang J, Kiba T, Park J, Kojima M, Do J, Kim KY, Kwon M, Endler A, Song W-Y, et al. (2014) Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc Natl Acad Sci USA 111: 7150–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E-J, Facchini PJ (2011) Tyrosine aminotransferase contributes to benzylisoquinoline alkaloid biosynthesis in opium poppy. Plant Physiol 157: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128 [DOI] [PubMed] [Google Scholar]
- Liscombe DK, Macleod BP, Loukanina N, Nandi OI, Facchini PJ (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66: 1374–1393 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Buron JD, Salomonsen B, Olsen CE, Hansen BG, Mortensen UH, Halkier BA (2012) Microbial production of indoleglucosinolate through enzyme engineering in a versatile yeast expression platform. Metab Eng 14: 104–111 [DOI] [PubMed] [Google Scholar]
- Minami H, Kim J-S, Ikezawa N, Takemura T, Katayama T, Kumagai H, Sato F (2008) Microbial production of plant benzylisoquinoline alkaloids. Proc Natl Acad Sci USA 105: 7393–7398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Shitan N, Sawada K, Van Montagu MCE, Inzé D, Rischer H, Goossens A, Oksman-Caldentey K-M, Moriyama Y, Yazaki K (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc Natl Acad Sci USA 106: 2447–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JS, Dastmalchi M, Li J, Chang L, Chen X, Hagel JM, Facchini PJ (2016) Plug-and-play benzylisoquinoline alkaloid biosynthetic gene discovery in engineered yeast. Methods Enzymol 575: 143–178 [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Matsumura E, Koyanagi T, Katayama T, Kawano N, Yoshimatsu K, Yamamoto K, Kumagai H, Sato F, Minami H (2016) Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat Commun 7: 10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Onoyovwe A, Hagel JM, Chen X, Khan MF, Schriemer DC, Facchini PJ (2013) Morphine biosynthesis in opium poppy involves two cell types: Sieve elements and laticifers. Plant Cell 25: 4110–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RME, Xu D, Foureau E, Teto Carqueijeiro MIS, Oudin A, Bernonville TD, Novak V, Burow M, Olsen CE, Jones DM, et al. (2017) An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat Plants 3: 16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reed JW, Hudlicky T (2015) The quest for a practical synthesis of morphine alkaloids and their derivatives by chemoenzymatic methods. Acc Chem Res 48: 674–687 [DOI] [PubMed] [Google Scholar]
- Roberts MF, McCarthy D, Kutchan TM, Coscia CJ (1983) Localization of enzymes and alkaloidal metabolites in Papaver latex. Arch Biochem Biophys 222: 599–609 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Chai G, Mehta S, Schick A (2018) Trends and economic drivers for United States naloxone pricing, January 2006 to February 2017. Addict Behav 86: 86–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsen B, Mortensen UH, Halkier BA (2014) USER-derived cloning methods and their primer design. Methods Mol Biol 1116: 59–72 [DOI] [PubMed] [Google Scholar]
- Samanani N, Liscombe DK, Facchini PJ (2004) Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J 40: 302–313 [DOI] [PubMed] [Google Scholar]
- Shitan N, Bazin I, Dan K, Obata K, Kigawa K, Ueda K, Sato F, Forestier C, Yazaki K (2003) Involvement of CjMDR1, a plant multidrug-resistance-type ATP-binding cassette protein, in alkaloid transport in Coptis japonica. Proc Natl Acad Sci USA 100: 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitan N, Dalmas F, Dan K, Kato N, Ueda K, Sato F, Forestier C, Yazaki K (2013) Characterization of Coptis japonica CjABCB2, an ATP-binding cassette protein involved in alkaloid transport. Phytochemistry 91: 109–116 [DOI] [PubMed] [Google Scholar]
- Shitan N, Minami S, Morita M, Hayashida M, Ito S, Takanashi K, Omote H, Moriyama Y, Sugiyama A, Goossens A, et al. (2014) Involvement of the leaf-specific multidrug and toxic compound extrusion (MATE) transporter Nt-JAT2 in vacuolar sequestration of nicotine in Nicotiana tabacum. PLoS One 9: e108789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitan N. (2016) Secondary metabolites in plants: Transport and self-tolerance mechanisms. Biosci Biotechnol Biochem 80: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Stanke M, Steinkamp R, Waack S, Morgenstern B (2004) AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res 32: W309–W312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N, Bürkle L, Pourcel L, Moulin M, Stolz J, Fitzpatrick TB (2013) Recycling of pyridoxine (vitamin B6) by PUP1 in Arabidopsis. Plant J 75: 40–52 [DOI] [PubMed] [Google Scholar]
- Takanashi K, Yamada Y, Sasaki T, Yamamoto Y, Sato F, Yazaki K (2017) A multidrug and toxic compound extrusion transporter mediates berberine accumulation into vacuoles in Coptis japonica. Phytochemistry 138: 76–82 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Arthanari H, Yang F, Pan S-J, Fan X, Breger J, Frueh DP, Gulshan K, Li DK, Mylonakis E, et al. (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452: 604–609 [DOI] [PubMed] [Google Scholar]
- Trenchard IJ, Siddiqui MS, Thodey K, Smolke CD (2015) De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab Eng 31: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker ME, Gardner JM, Vystavelova A, McBryde C, de Barros Lopes M, Jiranek V (2003) Application of the reuseable, KanMX selectable marker to industrial yeast: Construction and evaluation of heterothallic wine strains of Saccharomyces cerevisiae, possessing minimal foreign DNA sequences. FEMS Yeast Res 4: 339–347 [DOI] [PubMed] [Google Scholar]
- Weid M, Ziegler J, Kutchan TM (2004) The roles of latex and the vascular bundle in morphine biosynthesis in the opium poppy, Papaver somniferum. Proc Natl Acad Sci USA 101: 13957–13962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, Walker C, et al. (2012) A Papaver somniferum 10-gene clusterfor the synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang X (2016) Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng 37: 114–121 [DOI] [PubMed] [Google Scholar]
- Zhou K, Qiao K, Edgar S, Stephanopoulos G (2015) Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol 33: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulak KG, Khan MF, Alcantara J, Schriemer DC, Facchini PJ (2009) Plant defense responses in opium poppy cell cultures revealed by liquid chromatography-tandem mass spectrometry proteomics. Mol Cell Proteomics 8: 86–98 [DOI] [PubMed] [Google Scholar]
- Zürcher E, Liu J, di Donato M, Geisler M, Müller B (2016) Plant development regulated by cytokinin sinks. Science 353: 1027–1030 [DOI] [PubMed] [Google Scholar]