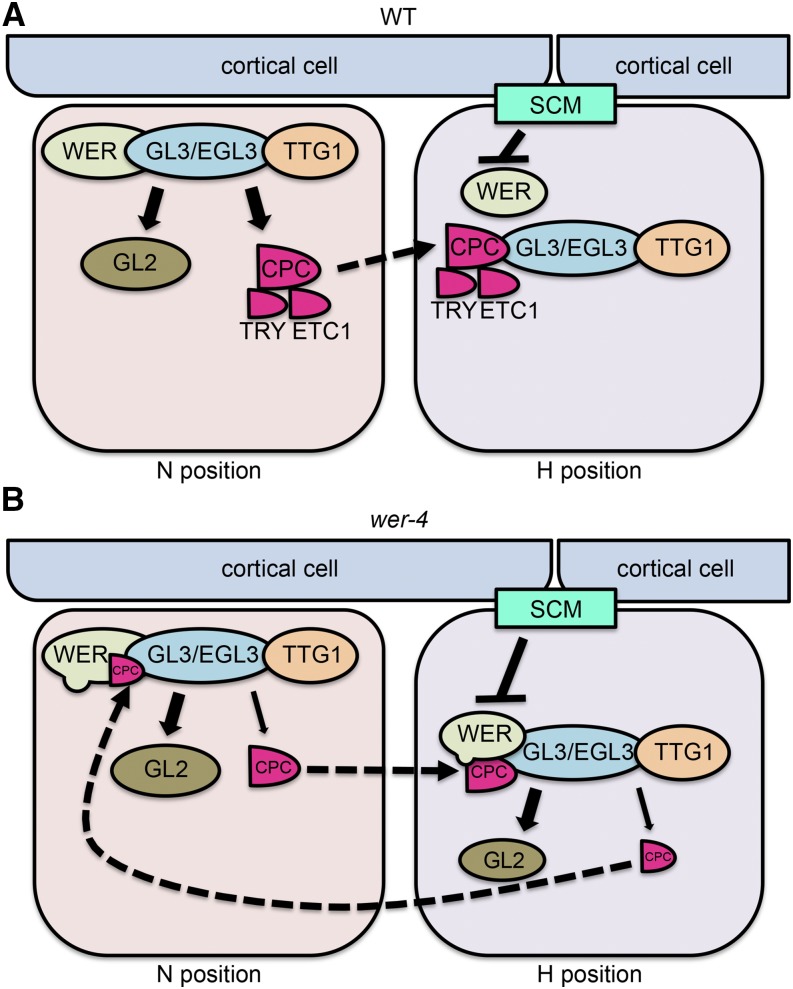
Figure 11.
Models for epidermal cell fate regulation in wild-type and wer-4 roots. The solid arrows (sharp or blunt) indicate transcriptional regulation. The dashed arrows indicate protein movement. A, In the wild-type (WT) root epidermis, the WER-GL3/EGL3-TTG1 complex preferentially accumulates in N-position cells and promotes the expression of GL2, CPC, TRY, and ETC1. The GL2 protein remains in N-position cells and inhibits root hair formation. The CPC, TRY, and ETC1 proteins move to adjacent H-position cells and compete with WER for GL3/EGL3 binding, allowing root hair formation. B, In the wer-4 mutant, the D105N residue substitution disrupts WER target gene transcription, largely abolishing TRY and ETC1 expression and reducing the expression of CPC relative to that of GL2. As a consequence, there is reduced competition for GL3/EGL3 binding and enhanced formation of the WER-GL3/EGL3-TTG1 complex in H-position cells. This triggers abnormal expression of GL2 and CPC in the H-position cells, leading to inappropriate CPC movement and accumulation in N-position cells as well as reduction of the WER-GL3/EGL3-TTG1 complex. Therefore, in the wer-4 root epidermis, the WER-GL3/EGL3-TTG1 complex accumulates abnormally in both H and N positions, leading to misspecification of cells in both positions.