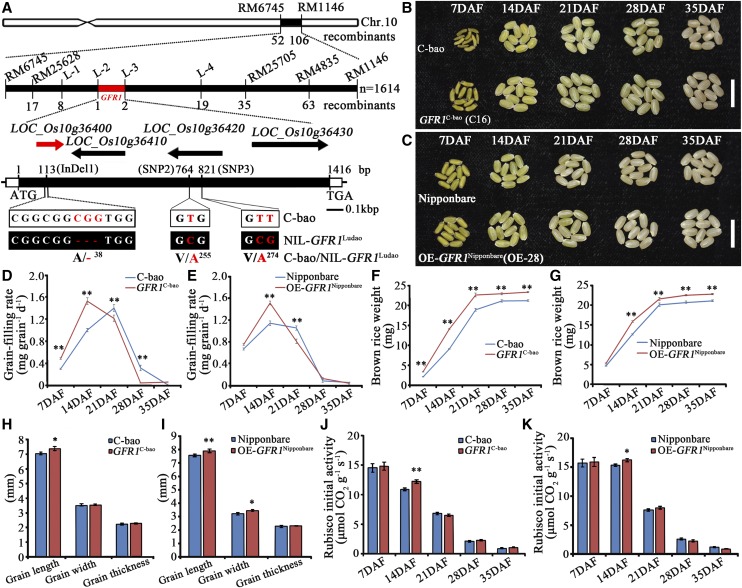
Figure 2.
Map-based cloning and identification of GFR1. A, GFR1 was finely mapped onto the long arm of chromosome 10. Molecular markers and numbers of recombinants are labeled above and below the filled bars, respectively. The candidate ORF1 (LOC_Os10g36400) is marked in red. The mutation sites of ORF1 are also shown. ATG and TGA represent the start and stop codons, respectively. B, Complementation identification of GFR1: morphological changes of brown rice grains of C-bao and C16 of the GFR1C-bao line (T3 transgenic plants) at five stages. Bar = 1 cm. C, Overexpression identification of GFR1: morphological changes of brown rice grains of Nipponbare and OE-GFR1Nipponbare (T3 transgenic plants) at five stages. Bar = 1 cm. D, Grain-filling rate changes in C-bao and GFR1C-bao. E, Grain-filling rate changes in Nipponbare and OE-28 of the OE-GFR1Nipponbare line. F, Brown rice weight changes of C-bao and GFR1C-bao. G, Brown rice weight changes of Nipponbare and OE-GFR1Nipponbare. H, Grain size of C-bao and GFR1C-bao. I, Grain size of Nipponbare and OE-GFR1Nipponbare. J, Initial Rubisco activity of C-bao and GFR1C-bao. K, Initial Rubisco activity of Nipponbare and OE-GFR1Nipponbare. The data are means ± sd (n = 3). Student’s t test was used for statistical analysis (*, P ≤ 0.05 and **, P ≤ 0.01).