Silencing of a light-signaling factor reveals developmental trade-offs between tomato plant growth, senescence, fruit yield, and nutritional quality.
Abstract
Plant development is highly dependent on the ability to perceive and cope with environmental changes. In this context, PIF proteins are key players in the cellular hub controlling responses to fluctuating light and temperature conditions. Reports in various plant species show that manipulation of the PIF4 level affects important agronomical traits. In tomato (Solanum lycopersicum), SlPIF1a and SlPIF3 regulate fruit nutraceutical composition. However, the wider role of this protein family, and the potential of their manipulation for the improvement of other traits, has not been explored. Here we report the effects of constitutive silencing of tomato SlPIF4 on whole-plant physiology and development. Ripening anticipation and higher carotenoid levels observed in SlPIF4-silenced fruits revealed a redundant role of SlPIF4 in the accumulation of nutraceutical compounds. Furthermore, silencing triggered a significant reduction in plant size, flowering, fruit yield, and fruit size. This phenotype was most likely caused by reduced auxin levels and altered carbon partitioning. Impaired thermomorphogenesis and delayed leaf senescence were also observed in silenced plants, highlighting the functional conservation of PIF4 homologs in angiosperms. Overall, this work improves our understanding of the role of PIF proteins—and light signaling—in metabolic and developmental processes that affect yield and composition of fleshy fruits.
Light is one of the most critical ambient factors controlling plant development, providing energy for photosynthesis and information about the constantly changing environment (McDonald, 2003). The ability to sense and adapt growth rhythms and metabolism to light conditions is paramount for plant survival (Kami et al., 2010). Phytochromes (PHYs) are red/far-red light photoreceptors, activated by light and deactivated by dark and high temperature (Wang and Deng, 2004; Jung et al., 2016; Legris et al., 2016). Upon light exposure, PHYs are translocated into the nucleus, where they interact with PHY-INTERACTING FACTORS (PIFs) and induce the degradation of these transcription factors. PIFs, in turn, act downstream of PHYs, repressing photomorphogenic responses in the dark. This interaction module regulates many developmental and physiological responses, such as deetiolation, growth, flowering, and senescence (Castillon et al., 2007; Leivar and Monte, 2014; Pham et al., 2018).
In tomato (Solanum lycopersicum), PHY-mediated light perception and PIF-dependent light signal transduction have been reported to regulate fruit development, nutritional quality, and ripening time (Azari et al., 2010; Cruz et al., 2018; Gramegna et al., 2019). For example, mutation and fruit-specific silencing of SlPHYA, SlPHYB1, and SlPHYB2 alter carbohydrate metabolism, sink activity, and carotenoid biosynthesis, ultimately affecting the nutritional composition of ripe fruits (Alba et al., 2000; Gupta et al., 2014; Bianchetti et al., 2018). In addition, the downregulation of PHY-signaling repressors, such as CONSTITUTIVE PHOTOMORPHOGENESIS 1 (SlCOP1), DEETIOLATED 1 (SlDET1), and SlPIF1a, has the opposite effect on ripe fruit pigmentation (Davuluri et al., 2004; Liu et al., 2004; Enfissi et al., 2010; Llorente et al., 2016). In line with these observations, SlPIF3 has been recently shown to repress tocopherol biosynthesis in tomato (Gramegna et al., 2019).
The study of functional conservation among PIFs has the potential to develop new tools for plant breeding, considering that these proteins control traits of agronomical importance. For instance, natural variation in Arabidopsis (Arabidopsis thaliana) AtPIF4 is associated with quantitative traits such as internode length, flowering time, and fruit setting (Brock et al., 2010). Additionally, variation of AtPIF4 gene expression is associated with heterosis. In this species, hybrid vigor correlates with increased expression of AtPIF4. It was proposed that this protein, at least in part, regulates hybrid vigor by inducing auxin biosynthesis and action, resulting in larger rosettes and increased biomass (Wang et al., 2017). Although manipulation of light signals bears a great potential to influence fruit yield, so far, PIF studies in tomato have been limited to impacts on isoprenoid metabolism in fruits (Llorente et al., 2016; Gramegna et al., 2019).
Among the multiple PIF-encoding genes in the tomato genome, SlPIF1a, SlPIF1b, SlPIF3, and SlPIF4 showed the highest expression levels in seedlings, leaves, and fruits (Rosado et al., 2016). Based on phylogenetic and transcriptional analyses, it has been proposed that SlPIF4 might have similar functions in the Arabidopsis orthologs AtPIF4 and AtPIF5 (Rosado et al., 2016). Therefore, SlPIF4 has the potential to regulate hypocotyl elongation, plant growth, flowering, and leaf senescence in response to light and temperature (Kunihiro et al., 2011; Kumar et al., 2012; Sun et al., 2012; Sakuraba et al., 2014; Xie et al., 2017). Here, we show that these functions are indeed shared, further strengthening the idea of the functional conservation of members of the PIF4 clade within angiosperms. Moreover, we demonstrate that manipulation of SlPIF4 levels has pleiotropic effects on tomato plant physiology, ultimately affecting yield and quality of the edible fruit.
RESULTS
Constitutive Silencing of Tomato SlPIF4
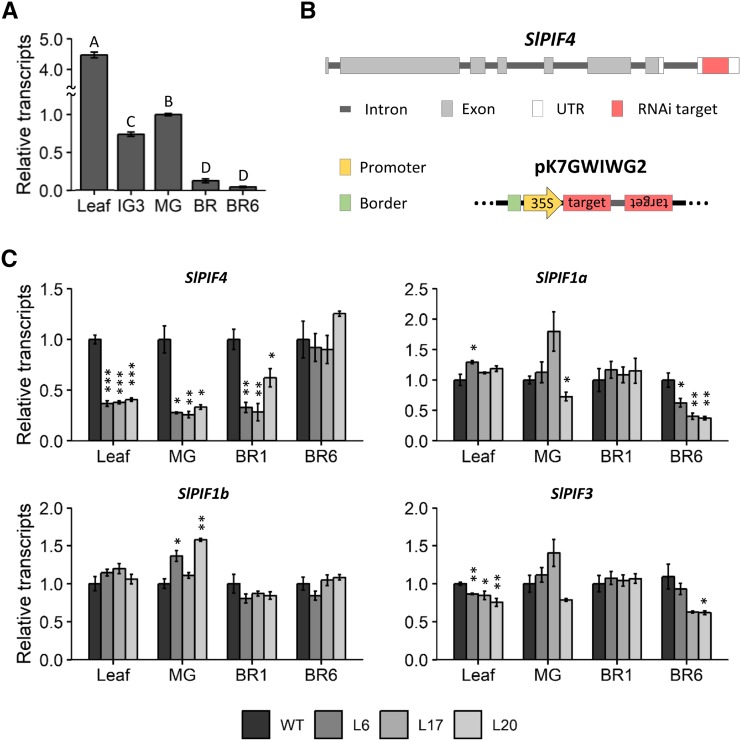
To investigate the role of PIF4 in tomato, we first investigated SlPIF4 expression under regular cultivation conditions (Fig. 1A). The highest mRNA levels were observed in leaves, whereas in fruits, SlPIF4 expression dramatically decreased upon ripening, confirming previous observations in detached fruits (Rosado et al., 2016). Considering this broad expression profile and the well-described role of AtPIF4 in several distinct physiological processes, we decided to generate constitutively SlPIF4-silenced lines by RNA interference (RNAi)-mediated knockdown. In order to avoid cosilencing of other SlPIFs, a sequence consisting of 180 bp of the 3′-untranslated region (UTR) of SlPIF4 was used to express a hairpin loop mRNA (Fig. 1B). Constitutive silencing with a reduction of at least 60% in transcript abundance in leaves and green fruits was confirmed by reverse-transcription quantitative PCR (RT-qPCR) analysis in three independent lines: 35S::SlPIF4-RNAi L6, 35S::SlPIF4-RNAi L17, and 35S::SlPIF4-RNAi L20, hereafter named L6, L17, and L20 (Fig. 1C). No cosilencing of SlPIF1a, SlPIF1b, or SlPIF3 was observed, although occasional reductions in expression were detected (Fig. 1C).
Figure 1.
Expression profile of SlPIF4 in wild-type plants and SlPIF genes in SlPIF4-silenced plants. A, Transcript profile of SlPIF4 in wild-type (WT) plants. B, SlPIF4 gene structure showing the RNAi target sequence on the 3′ UTR in red. Also shown is the construct used for silencing in the pK7GWIWG2 vector. C, mRNA abundance of SlPIF genes in SlPIF4-silenced plants. Data shown are the mean ± se of at least three biological replicates (each composed of four fruits or two leaves) normalized against the mature-green (MG) stage (A) or the wild-type control (C). Significant differences between samples (A) and relative to the wild-type control (C) are denoted by uppercase letters (ANOVA followed by Fisher’s lsd test) and asterisks (two-tailed t test; *P < 0.05; **P < 0.01; ***P < 0.001), respectively. Leaf, the fourth leaf of 90 day-old plants; IG3, immature-green; BR, breaker stage; BR1, one day after BR stage; BR6: 6 days after BR stage; L6, 35S::SlPIF4-RNAi L6; L17, 35S::SlPIF4-RNAi L17; L20, 35S::SlPIF4-RNAi L20.
SlPIF4 Regulates Fruit Ripening and Quality
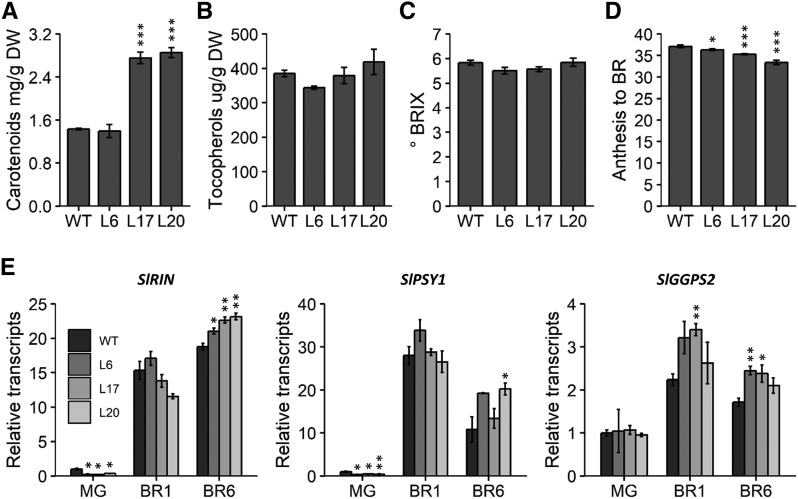
Two previous studies in tomato (Llorente et al., 2016; Gramegna et al., 2019) showed that SlPIF1a and SlPIF3 had roles in inhibiting the accumulation of nutraceutical compounds, in particular carotenoids and tocopherols, respectively, during ripening. To test whether this is a conserved function among tomato PIFs, we evaluated the levels of these isoprenoid-derived compounds, as well as total soluble solids content (refractive index, °BRIX), in ripe fruits (12 d after breaker [BR] stage, i.e. fully red ripe fruits). Carotenoid levels were up to 2-fold higher than in the wild-type counterparts in two of the transgenic lines (L17 and L20; Fig. 2A). In contrast, no significant changes in tocopherol and °BRIX were detected between the transgenic and wild-type fruits (Fig. 2, B and C).
Figure 2.
SlPIF4 silencing affects tomato fruit quality. A to C, Carotenoid (A), tocopherol (B), and °BRIX (C) in ripe fruits (12 d post breaker stage [BR]). D, Ripening time from anthesis to BR stage. E, mRNA abundance relative to wild-type (WT) MG of differentially expressed genes involved in carotenogenesis. Values represent means ± se of at least three biological replicates, each composed of at least four fruits (A, B, and E), 10 individual fruits (C), or 90 individual fruits (D). Significant differences relative to the wild-type control are denoted by asterisks (two-tailed t test; *P < 0.05; **P < 0.01; ***P < 0.001). BR1-12, 1-12 days after BR stage; RIN, RIPENING INHIBITOR; PSY1, PHYTOENE SYNTHASE; GGPPS2, GERANYL GERANYL DIPHOSPHATE SYNTHASE; L6, 35S::SlPIF4-RNAi L6; L17, 35S::SlPIF4-RNAi L17; L20, 35S::SlPIF4-RNAi L20.
Interestingly, SlPIF4-silenced fruits not only accumulated more carotenoids but also ripened faster than control fruits considering the time from anthesis to the BR stage (Fig. 2D). We further confirmed this phenotype by analyzing colorimetric parameters of detached fruits throughout ripening (Supplemental Fig. S1). In accordance with the observed advance in ripening, the color change was initially faster from the BR to BR2 stage (2 d after BR) in fruits from L20 homozygous SlPIF4-silenced plants. In line with their higher lycopene content, SlPIF4-silenced ripe fruits showed more intense red color in comparison to the wild type (Supplemental Table S1). Higher transcript abundance of the ripening master regulator RIPENING INHIBITOR (SlRIN) and the key genes involved in carotenoid biosynthesis, namely GERANYL GERANYL DIPHOSPHATE SYNTHASE (SlGGPS2) and PHYTOENE SYNTHASE (SlPSY1), detected in the transgenic lines explains these phenotypes, at least in part, and suggests a role for SlPIF4 in the regulation of fruit ripening and carotenogenesis (Fig. 2E).
SlPIF4 Silencing Impacts Flowering and Fruit Production
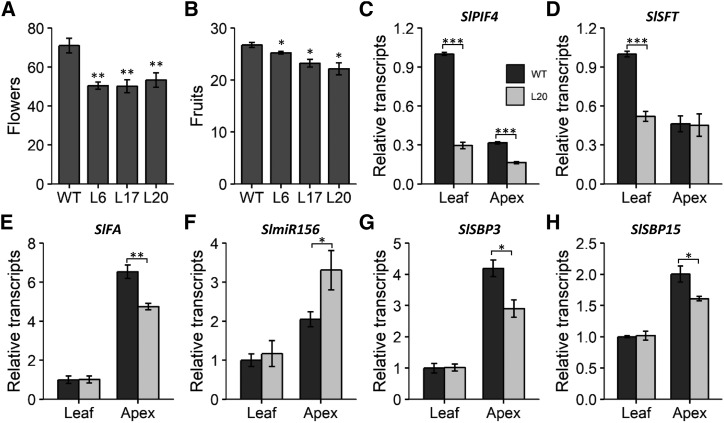
Flowering control by AtPIF4 has been extensively reported in Arabidopsis (Brock et al., 2010; Kumar et al., 2012; Thines et al., 2014; Galvão et al., 2015; Seaton et al., 2015; Fernández et al., 2016). In this species, AtPIF4 induces the florigen FLOWERING LOCUS T (AtFT) directly by binding to its promoter and indirectly by repressing microRNA156 (AtmiR156) expression. Flowering in tomato is regulated similarly; SlmiR156 represses the expression of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 (SlSPB3) and SlSBP15 in both apex and leaf. In turn, these proteins induce the expression of SINGLE FLOWER TRUSS (SlSFT), the AtFT ortholog, in leaves and FALSIFLORA (SlFA) in shoot apices (Silva et al., 2019). SlSFT protein is translocated to the apex and together with FA induces flowering (Molinero-Rosales et al., 2004). However, the role of SlPIF4 in this regulatory network has not been addressed yet. Thus, we tested whether flowering was also affected by reduced SlPIF4 expression in tomato. In silenced lines, a significant reduction in flower number was observed, which was reflected by reduced fruit production in 18-week-old plants (Fig. 3, A and B). Interestingly, individual transgenic trusses produced fewer flowers than in the wild type, as addressed for the two first flowering trusses (Supplemental Fig. S2A). No changes in flowering time were observed between the studied genotypes when either the number of leaves until the first truss or the number of days until the anthesis of the first flower per plant were scored (Supplemental Fig. S2, B and C). In order to understand the molecular mechanism behind this phenotype, the miR156-SPB-SFT/FA module that regulates flowering in tomato (Silva et al., 2019) was profiled in leaves and shoot apices harvested from wild-type and L20 30-d-old young plants (Fig. 3, C–H). SlPIF4 was shown to be under-expressed in the apex as compared to leaves, and silencing was confirmed in both organs. Downregulation of SlSFT and SlFA florigens was observed in leaves and apices, respectively, of SlPIF4-silenced plants. Moreover, the abundance of miR156 increased in the apex of transgenic plants, which negatively correlated with its targets SlSBP3 and SlSBP15 in the same organ. These data demonstrated that SlPIF4 regulates flowering in tomato, reinforcing the hypothesis that members of the PIF4 clade play a conserved role in angiosperms.
Figure 3.
SlPIF4 silencing affects plant development and fruit yield. A and B, Total flower and fruit number produced by T2 18-week-old plants. Values represent means ± se for at least six different plants. C to H, Transcript profile of flowering genes in 30-d-old T4 plants. Values represent means ± se for three biological replicates composed of two leaves or apices. Significant differences relative to the wild-type (WT) control are denoted by asterisks (two-tailed t test; *P < 0.05; **P < 0.01; ***P < 0.001). L6, 35S::SlPIF4-RNAi L6; L17, 35S::SlPIF4-RNAi L17; L20, 35S::SlPIF4-RNAi L20; PIF4, PHYTOCHROME INTERACTING FACTOR 4; SFT, SINGLE FLOWER TRUSS; FA, FALSIFLORA; SBP3 and 15, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 and 15; miR156, microRNA 156.
SlPIF4 Silencing Impacts Vegetative Growth and Fruit Size
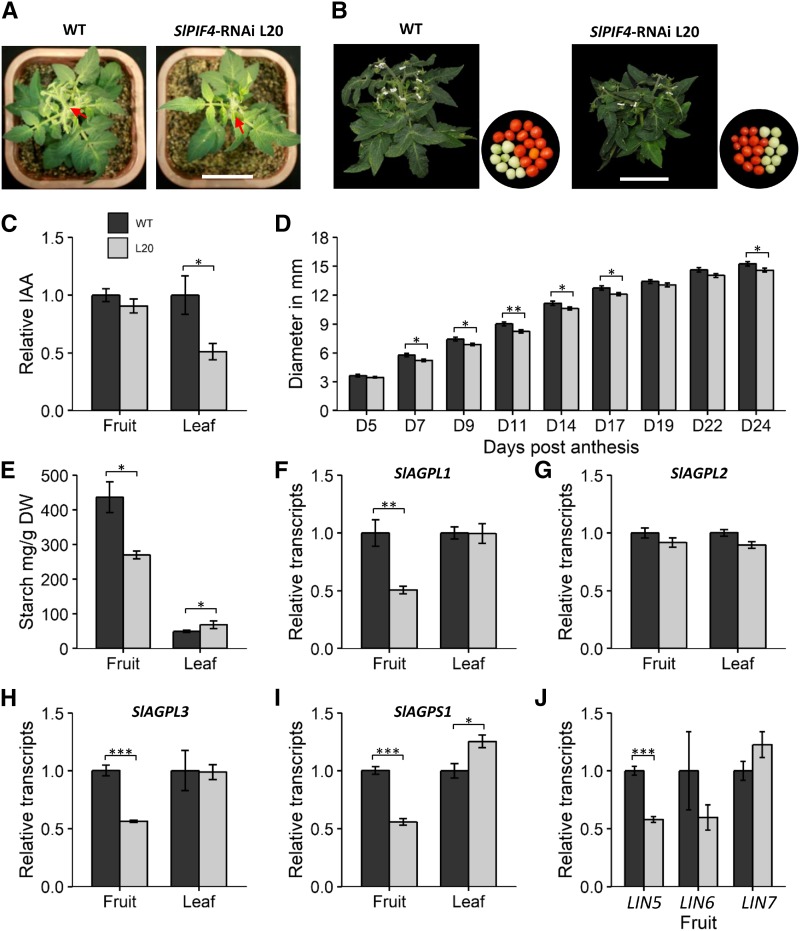
To better understand to what extent constitutive silencing affected fruit yield, we compared different growth and production parameters in wild-type and L20 homozygous plants. At an early age (5 weeks old), SlPIF4-silenced plants were visually smaller than wild-type plants (Fig. 4A). Differences in size were accentuated during the life cycle, and 18-week-old plants showed clear differences in size and fruit production (Fig. 4B), with SlPIF4-silencing causing an observed 15% reduction in vegetative weight and 23% reduction in fruit weight, accounting for a total reduction of 21% in plant aerial mass (Supplemental Table S2). Interestingly, fruit production was affected beyond number, as SlPIF4-silenced individual red fruits were smaller in mass and diameter compared to the wild type (Fig. 4B; Supplemental Table S2). These developmental differences could not be attributed to altered carbon assimilation rates since no alterations in photosynthesis were detected (Supplemental Table S3). Instead, the observed phenotype is most likely caused by the reduction of auxin levels (Fig. 4C) and aggravated by the reduction in overall carbon assimilation due to lowered leaf area (Fig. 4A) and number (Supplemental Table S2).
Figure 4.
SlPIF4 silencing affects growth and source-sink relationship. A, Representative 5-week-old wild-type and SlPIF4-silenced plants, showing developmental delay in the SlPIF4-silenced line. The red arrow indicates the first inflorescence. Bar = 5 cm. B, Representative 15-week-old plants, showing the differences in size and fruit production in the SlPIF4-silenced plants relative to the wild type. Bar = 10 cm. C and E to J, Relative auxin level (C), starch content (E), and transcript profile of starch biosynthetic and cell wall invertase genes (F to J) in immature-green fruits and source leaves. Values represent means ± se of at least three biological replicates composed of two leaves or four fruits. D, Size differences between wild-type and SlPIF4-silenced fruits of 5–24 days post anthesis (D5 to D24). Values represent means ± se of at least 20 individual fruits. In C to J, significant differences relative to the wild-type (WT) control are denoted by asterisks (two-tailed t test; *P < 0.05; **P < 0.01; ***P < 0.001). L20, homozygous T4 35S::SlPIF4-RNAi L20; AGPL1-3, ADP-GLUCOSE PYROPHOSPHORYLASE LARGE SUBUNIT 1-3; AGPS1, ADP-GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1; LIN5-7, LYCOPERSICUM INVERTASE 5-7.
On the other hand, although differences in size appeared early in fruit development (Fig. 4D), impaired fruit growth in SlPIF4-silenced plants could not be directly explained by the reduction of auxin levels, since no differences were detected in immature fruits (Fig. 4C). Interestingly, carbohydrate profiling revealed a shift in sugar partitioning in the silenced lines. Whereas no changes in soluble sugars were observed (Supplemental Table S4), starch was accumulated at higher levels in leaves and reduced in fruits of the L20 homozygous transgenic plants (Fig. 4E). These observations were in accordance with the expression profile of ADP-GLC PYROPHOSPHORYLASE (AGPase) large- and small-subunit-encoding genes (SlAGPL1, SlAGPL2, SlAGPL3, and SlAGPS1) involved in starch biosynthesis in both organs (Fig. 4, F–I). Additionally, expression of the flower- and fruit-specific invertase encoding gene LYCOPERSICUM INVERTASE 5 (SlLIN5; Fridman et al., 2004) was reduced in transgenic fruits (Fig. 4J), which could be indicative of reduced sink strength caused by SlPIF4 silencing. Thus, these observations further support the functional conservation of PIF4 in regulating plant growth and auxin biosynthesis, but they also illustrate a new role for SlPIF4 protein in fruit yield.
SlPIF4 Participates in Thermomorphogenesis
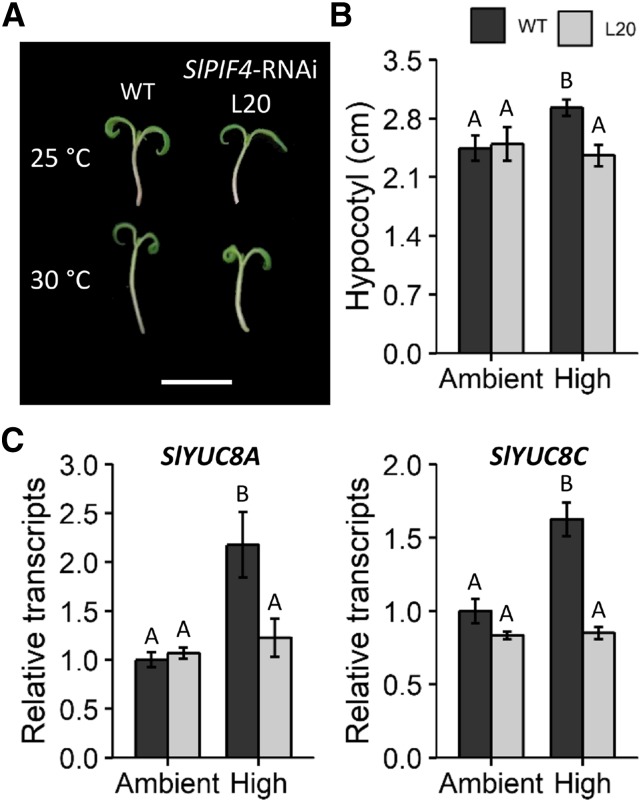
Beyond light, temperature is a key factor regulating plant growth and development (Kami et al., 2010; Quint et al., 2016), and many studies performed in Arabidopsis have placed AtPIF4 as an integrator of light and temperature responses (Franklin et al., 2011; Sun et al., 2012; Gangappa and Kumar, 2017). To address whether tomato SlPIF4 also participates in temperature perception, hypocotyl elongation was analyzed in seedlings maintained for 3 d in either ambient- (25°C) or high-temperature (30°C) conditions under a day-neutral photoperiod. Only wild-type seedlings responded to the treatment and showed longer hypocotyls at 30°C, whereas the hypocotyl length of SlPIF4-silenced seedlings remained unchanged (Fig. 5, A and B). Expression analysis revealed the up-regulation of YUCCA FLAVIN MONOOXYGENASES, SlYUC8A, and SlYUC8C, in wild-type seedlings in high-temperature conditions compared to that in SlPIF4-silenced seedlings (Fig. 5C), suggesting that the observed high temperature-associated elongation is the consequence of auxin biosynthesis enhancement, demonstrating SlPIF4 involvement in temperature responsiveness.
Figure 5.
SlPIF4 participates in high temperature-induced hypocotyl elongation. A, Composite image of representative seedlings depicting differences in size in response to temperature. Bar = 2 cm. B, Hypocotyl length. C, Relative transcript profile of auxin biosynthetic genes. Data shown are the means ± se of at least 14 seedlings (B) or three biological replicates composed of five seedlings each (C). Ambient and high temperatures were 25°C and 30°C, respectively. Significant differences relative to the wild-type (WT) control are denoted by uppercase letters (ANOVA followed by Fisher’s lsd test). L20, T4 homozygous 35S::SlPIF4-RNAi L20; YUC8A-C, YUCCA FLAVIN MONOOXYGENASE 8A-C.
Tomato PIF4 Promotes Age-Induced Leaf Senescence
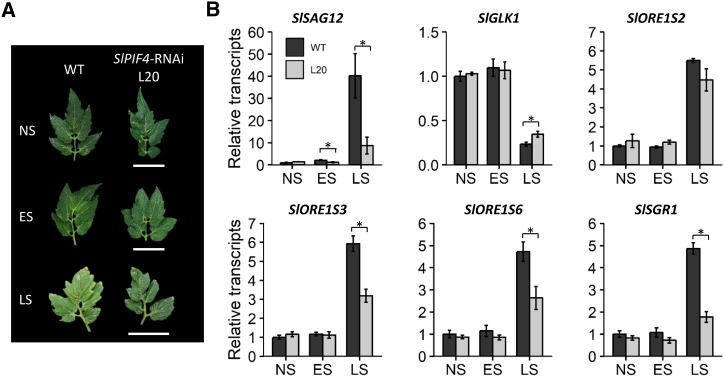
In Arabidopsis, AtPIF4 and AtPIF5 promote both age- and dark-induced senescence by activating ORESARA 1 (ORE1) transcription factors, genes involved in chlorophyll breakdown, such as STAYGREEN (SGR), and repressing chloroplast maintainer GOLDEN2-LIKE 1 (GLK1; Sakuraba et al., 2014; Song et al., 2014; Zhang et al., 2015). Although the same downstream effectors are involved in leaf senescence in tomato (Lira et al., 2017), the role of SlPIF4 in this signaling pathway has not been addressed so far. To investigate its involvement, leaves without any signs of senescence (nonsenescent), with initial yellowing (early senescent), and with advanced yellowing (late senescent) were harvested from wild-type plants. Leaves from corresponding phytomeres from SlPIF4-silenced plants were also collected. Visually, silenced leaves remained greener than control leaves (Fig. 6A), suggesting that senescence was delayed in these plants. Lowered expression of the senescence marker SENESCENCE ASSOCIATED GENE12 (SlSAG12) confirmed this hypothesis. Also, higher expression of chloroplast maintainer SlGLK1 and reduced levels of senescence-associated transcription factors SlORE1S23 and SlORE1S26, as well as SlSGR1, possibly contributed to the observed staygreen phenotype (Fig. 6B). Thus, similar to that described in Arabidopsis, SlPIF4 participates in the senescence-inducing pathway.
Figure 6.
SlPIF4 silencing delays age-induced leaf senescence. A, Composite image of representative nonsenescent (NS, phytomer 6), early-senescent (ES, phytomer 4), and late-senescent (LS, phytomers 1 and 2) leaves from wild-type and SlPIF4-silenced 18-week-old plants. Bars = 5 cm. B, Transcript profile of senescence-related genes. Data shown are means ± se of at least three individual leaves. Significant differences with wild-type (WT) controls in two-tailed t test: *P < 0.05. L20, 35S::SlPIF4-RNAi L20; SAG12, SENESCENCE ASSOCIATED GENE 12; GLK1, GOLDEN-2 LIKE 1; ORE1S02-6, ORESARA 1 LIKE 2-6; SGR1, STAY-GREEN 1.
DISCUSSION
Although PIF genes have been extensively studied in Arabidopsis for over 20 years (Ni et al., 1998), only recently have they been identified in tomato (Rosado et al., 2016). The only SlPIFs studied so far are SlPIF1a and SlPIF3, which have been demonstrated to regulate fruit nutraceutical value (Llorente et al., 2016; Gramegna et al., 2019). With the aim of expanding our knowledge on the potential biotechnological use of the PIF protein family in crop species, here we comprehensively characterized SlPIF4-silenced tomato plants taking into account the role of PIF4 in flowering time and fruit setting in Arabidopsis (Brock et al., 2010) and, more recently, in biomass production in switchgrass (Panicum virgatum; Yan et al., 2018).
The silencing of SlPIF4 plants resulted in downregulation of SlPIF1a and SlPIF3 exclusively in red fruits and leaves, respectively (Fig. 1C). However, this downregulation cannot be attributed directly to expression of the silencing construct since it was not observed in other stages. Additionally, since the fragment used for the silencing construct was from the SlPIF4 3′UTR sequence (Fig. 1B) and an off-target analysis was carefully performed, it is more likely that differential expression of SlPIF1a and SlPIF3 is a side effect of SlPIF4 silencing rather than cosilencing. This is in line with the regulatory network proposed for Arabidopsis, in which PIFs regulate each other at the transcriptional level (Leivar and Monte, 2014).
Interestingly, although SlPIF4 is poorly expressed in wild-type ripening fruits (Fig. 1A), the silencing of this gene had a considerable effect on the ripening process (Fig. 2; Supplemental Fig. S1), uncovering a role for tomato PIFs in regulating ripening time. The late increase in expression of the master ripening regulator SlRIN observed in BR6 transgenic fruits cannot explain the ripening advance but may contribute to carotenoid accumulation and induction of SlPSY1 in ripe fruits (Fig. 2; Fujisawa et al., 2011). The upregulation of expression of SlPSY1 and SlGGPS2, whose encoded enzymes act upstream in carotenogenesis, explains the accumulation of all carotenoid forms in ripe fruits (Supplemental Table S1). It is unlikely, though, that SlPIF4 directly regulates carotenogenesis, because no differential carotenoid accumulation occurred in SlPIF4-silenced MG fruits, when SlPIF4 is normally highly expressed (Fig. 1A; Supplemental Table S1,). Previous work showed that changes in pigment composition during ripening alter the quality of the light filtered through the fruit pericarp, increasing the relative red/far-red ratio (Llorente et al., 2016). As a consequence, PIF degradation increases, enhancing SlPSY1 expression and carotenoid accumulation. The reduced SlPIF1a mRNA levels, which inversely correlate with SlPSY1 expression in transgenic BR6 fruits, might additionally contribute to the observed phenotypes at this stage (Figs. 1C and 2). Since no changes in tocopherols were detected in SlPIF4-silenced fruits (Fig. 2B), it is possible that only SlPIF3 participates in the regulation of tocopherol biosynthesis, as previously proposed (Gramegna et al., 2019). Nevertheless, the presented data demonstrate a functional convergence of tomato PIFs in regulating fruit nutritional quality (Llorente et al., 2016; Bianchetti et al., 2018; Gramegna et al., 2019), and suggest a unique role of SlPIF4 in the regulation of ripening time and progression, which should be further dissected in future research.
Manipulation of flowering-related traits is a key strategy to improve fruit yield in tomato (Krieger et al., 2010). Indeed, SlPIF4 silencing had an impact on fruit production derived from lowered flower number (Fig. 3; Supplemental Table S2). Considering that altered PIF4 levels in other species, such as Arabidopsis, rice (Oryza sativa), and maize (Zea mays), also affect flowering, our data agree with the notion of a conserved function of PIF4 angiosperm homologs (Kumar et al., 2012; Kudo et al., 2017; Shi et al., 2018). Accordingly, both tomato and Arabidopsis PIF4 induce flowering via the miR156-SPB-florigen (SlSFT or AtFT) module (Fig. 3; Xie et al., 2017). However, SlPIF4 silencing only affected flower number (Fig. 3), not flowering time (Supplemental Fig. S2, B and C), contrary to findings in the Arabidopsis pif4 mutant (Brock et al., 2010; Thines et al., 2014; Galvão et al., 2015). We attribute this difference to two facts. First, these species have different requirements for flowering: whereas domesticated tomato is a day-neutral species (Soyk et al., 2017), Arabidopsis is a long-day plant (Cho et al., 2017); therefore, alterations of light-sensing in tomato are not expected to affect flowering time. Second, whereas mutations in SlSFT affect flowering time by changing the rate of truss production (Molinero-Rosales et al., 2004), reduction of SlSFT transcripts, as observed in SlPIF4-silenced plants, are expected to have only mild effects on flowering and would not necessarily affect flowering time. However, downregulation of SlSFT caused a reduction in the number of flowers produced per truss (Supplemental Fig. S2A), in agreement with observations in sft mutants displaying altered inflorescence development (Molinero-Rosales et al., 2004). On the other hand, it is possible that manipulation of SlPIF4 in wild tomato species would bear different results, since wild tomatoes flower earlier under short days (Soyk et al., 2017). Loss of photoperiod sensitivity in domesticated varieties has been associated with mutations at the SELF PRUNING 5G (SP5G) locus, an antiflorigen. Such mutations reduce the expression of this gene under long-day conditions, therefore attenuating the photoperiodic response (Cao et al., 2016; Soyk et al., 2017; Zhang et al., 2018). Interestingly, SlPHYB1 regulates SP5G expression (Cao et al., 2018), which makes this gene a likely target of regulation by SlPIF4 and reinforces the idea of a possible effect of SlPIF4 on flowering time in wild species. Recent work in tomato has shown that the tomato DELLA, PROCERA, which is involved in gibberellin signaling, induces flowering via the miR156-SPB-SFT module (Silva et al., 2019). However, in Arabidopsis, DELLA proteins are flowering repressors, which inhibit the activity of AtPIF4 (de Lucas et al., 2008; Xu et al., 2016). In this sense, investigation of tomato PROCERA-PIF4 interaction could reveal new layers of species-specific regulation of flowering.
In SlPIF4-silenced plants, lowered fruit number was accompanied by a reduction in ripe fruit size (Fig. 2; Supplemental Table S2), revealing a critical function of SlPIF4 in determining tomato yield. This phenotype was attributed to both impaired vegetative growth and altered source-sink relationship (Fig. 4). These observations are in agreement with a previous study in tomato that showed the importance of fruit-localized PHY for sugar partitioning and sink strength (Bianchetti et al., 2018). Fruit-specific SlPHYA or SlPHYB2 silencing cause overaccumulation of starch in immature fruits, which correlates with upregulation of genes involved in starch biosynthesis and cell wall invertases, such as SlLIN5. Since silencing of SlPIF4 had the opposite effect on starch synthesis and sink strength (Fig. 4), we propose that SlPHYA and SlPHYB2 regulate these processes via SlPIF4. A link between sugars and PIFs has been reported previously in Arabidopsis. In this species, sugars induce the expression of AtPIF4 and AtPIF5, coupling growth to carbon availability (Lilley et al., 2012; Sairanen et al., 2012). Here we show that SlPIF4 controls sugar partitioning and thus regulates photoassimilate exportation from source leaves toward sink organs, uncovering a further role for this transcription factor.
The results showed here for adult plants (Fig. 4) and seedlings (Fig. 5) suggest that SlPIF4 regulates growth by inducing auxin biosynthesis, which is in agreement with observations in Arabidopsis (Nozue et al., 2007; Niwa et al., 2009; Kunihiro et al., 2011; Nieto et al., 2015), rice (Todaka et al., 2012), and maize (Shi et al., 2018) that identified PIF4 as a growth regulator. Additionally, loss of temperature responsiveness in SlPIF4-silenced seedlings reveals another conserved feature for members of the PIF4 clade in angiosperms (Fig. 5; Koini et al., 2009). Recent works have placed PHYTOCHROME B (PHYB) as an integrator of light and temperature perception and PIF4 as a key protein in mediating the responses to both signals in Arabidopsis (Jung et al., 2016; Legris et al., 2016). In this context, future studies investigating interactions between SlPIF4, SlPHYB1, and SlPHYB2 will add invaluable information to understanding photo- and thermomorphogenesis in tomato.
Finally, SlPIF4-silenced plants displayed a delay in age-induced leaf senescence (Fig. 6), explained by the downregulation of SlORE1 transcription factor-encoding genes. These senescence-associated proteins negatively regulate chloroplast maintainer SlGLK1 and upregulate the expression of chlorophyll degradation enzymes, such as SlSGR (Lira et al., 2017), resulting in the staygreen phenotype observed in the old transgenic leaves (Fig. 6). It has been demonstrated that in Arabidopsis, AtPIF4 regulates SGR, ORE, and GLK by directly binding to specific motifs in their promoter regions (Sakuraba et al., 2014; Song et al., 2014; Zhang et al., 2015). Moreover, overexpression of maize ZmPIF4 and ZmPIF5 accelerates leaf senescence in Arabidopsis (Shi et al., 2018). These data strongly support that PIF4 is functionally conserved among angiosperms, as previously suggested (Rosado et al., 2016).
In summary, besides the functions previously described for PIF4, our results present additional roles for this protein in the regulation of sugar partitioning, fruit production, and ripening. Overall, the pleiotropic effects observed in SlPIF4-silenced plants not only highlight the importance of PIFs in plant development, but also suggest that manipulation of light signaling is an efficient strategy to improve tomato yield and quality.
MATERIALS AND METHODS
Plant Material, Growth Conditions, and Sampling
SlPIF4-silenced lines were generated by constitutively expressing an intron-spliced hairpin RNA construct containing a 180-bp fragment of the 3′ UTR region of SlPIF4 locus (Solyc07g043580). To avoid off-target effects, the construct was designed to have minimal complementarity with other genes, especially other SlPIFs, and then the sense/antisense fragment was used as a query for a BLAST search against the Sol Genomics Network database (www.solgenomics.net). The fragment was amplified from complementary DNA with the primers listed in Supplemental Table S5, cloned into pK7GWIWG2(I) (Karimi et al., 2002), and introduced into tomato (Solanum lycopersicum) cv Micro-Tom harboring the wild-type SlGLK2 allele (Carvalho et al., 2011) via Agrobacterium-mediated transformation according to Pino et al. (2010), with modifications described in Bianchetti et al. (2018). Despite its partial impairment in brassinosteroid biosynthesis due to the weak mutation d, Micro-Tom cultivar has been extensively demonstrated to represent a convenient and adequate model system to study fruit biology (Campos et al., 2010). The presence of the transgene was confirmed by PCR using the primers 35S forward and RNAi-specific reverse (Supplemental Table S5). After silencing verification by RT-qPCR analysis, three transgenic lines with a reduction of ∼60% in SlPIF4 mRNA level were selected for further analyses: 35S::SlPIF4-RNAi L6, 35S::SlPIF4-RNAi L17, and 35S::SlPIF4-RNAi L20. Two different generations of silenced plants were used in this work: L6, L17, and L20 segregating lines in T2, and the L20 homozygous line in T4.
Plants were grown in 6-l pots containing a 1:1 mixture of commercial substrate (Plantmax HT) and vermiculite, supplemented with 1 g L−1 of NPK 10:10:10, 4 g L−1 of dolomite limestone, and 2 g L−1 Yoorin Master (Yoorin Fertilizantes). Cultivation was carried out in a growth chamber with controlled light and temperature conditions (250 µmol m−2 s−1, 12 h/12 h photoperiod, 25 ± 2°C) and manual irrigation. For senescence analysis, T4 L20 plants were grown in a greenhouse (25 ± 2°C) with natural light conditions. Two T2 experiments were set: one for nondestructive total flower and fruit number, and another one for fruit harvesting. Third leaves completely expanded from 90-d-old plants were collected. Fruit pericarp was sampled at the MG, BR1 (1 d after BR), BR6, and BR12 stages. All further experiments were performed with T4 homozygous L20 plants. For colorimetric parameter measurement, fruits at the MG stage were harvested, placed in a 0.5-l sealed transparent vessel, and continuously flushed with ethylene-free humidified air (∼1 L min−1) at 12 h/12 h photoperiod conditions, 25 ± 2°C, and air relative humidity at 80% ± 5%. Colorimetric parameters were scored at MG, BR, BR1, BR2, BR3, BR6, and BR12 stages. For flowering experiments, the third leaf and shoot apex of 30-d-old plants were harvested. For growth and source-sink relationship analyses, the sixth leaves and immature green fruits from 12-week-old plants were collected. Yield was scored in 15-week-old plants. Ripe tomato size parameters were determined using Tomato Analyzer software (Rodriguéz et al., 2010). Immature fruit diameter was measured with digital calipers. Hypocotyl lengths were obtained from images analyzed in ImageJ software (https://imagej.nih.gov/ij/). Temperature experiments were performed in vitro. For that, seeds were sown in Murashige and Skoog growth media (Murashige and Skoog, 1962) and kept in the dark for 5 d at 25 ± 2°C; then, seedlings were transferred to 12 h/12 h photoperiod conditions under either 25 ± 2°C or 30 ± 2°C for 3 d. All samples were harvested ∼4–6 h after lights were turned on, immediately frozen in liquid nitrogen, and stored at −80°C until use.
RT-qPCR
RNA extraction, complementary DNA synthesis, and qPCR were performed as described by Quadrana et al. (2013) following Minimum Information for Publication of Quantitative Real-Time PCR Experiments guidelines (Bustin et al., 2009). Stem-loop pulse reverse transcription for SlmiR156 quantification was performed as described previously by Varkonyi-Gasic et al. (2007). qPCR was carried out in the QuantStudio 6 Flex Real-Time PCR system (Applied Biosystems) using 2× Power SYBR Green Master Mix (Life Technologies) in a 10-µL final volume. Quantitation cycle values and PCR efficiencies were obtained from absolute fluorescence data analyzed in the LinRegPCR software package (Ruijter et al., 2009). Expression values were normalized with TIP41 and EXPRESSED reference genes (Expósito-Rodríguez et al., 2008). All primers and accession numbers can be found in Supplemental Table S5.
Fruit Color, Carotenoid, Tocopherol, and °BRIX Determination
Fruit color and intensity (hue angle and chroma) were determined using a Konica Minolta CR-400 colorimeter as described in Su et al. (2015). Carotenoid extraction was carried out as described in Bianchetti et al. (2018) with modifications. Briefly, 20 mg of freeze-dried fruit pericarps were homogenized sequentially with 100 µL of saturated NaCl, 200 µL of dichloromethane, and 1 mL of hexane:diethyl ether (1:1, v/v). Supernatant was collected after centrifugation and pellets were re-extracted an additional three times with 500 µL hexane:diethyl ether mixture. Supernatant fractions were combined, vacuum-dried, suspended in 200 µL of acetonitrile, and filtered through a 0.45-µm membrane. Tocopherol extraction was performed as described by Lira et al. (2016). Briefly, 25 mg of freeze-dried fruit pericarps were homogenized sequentially in 1.5 mL methanol, 1.5 mL chloroform, and 2.5 mL Tris NaCl (Tris 50 mm, pH 7.5, and 1 m NaCl) solution. Following centrifugation, the organic fraction was collected and samples were re-extracted in 2 mL chloroform. Fractions were combined, then 3 mL of combined samples was vacuum dried, suspended in 200 µL of hexane:tert-butyl methyl ether (90:10, v/v), and filtered through a 0.45-µm membrane. Carotenoids and tocopherol levels were determined by HPLC in an Agilent 1100, as described in Lira et al. (2017). Total soluble sugars measured as °BRIX were determined in ripe (BR12) fruits as follows. Fresh pericarp tissue was homogenized with metallic beads and briefly spun. °BRIX of the resulting juice was measured in a portable digital refractometer NR151 (J.P. Selecta).
Hormone Analysis
Indole-3-acetic acid (IAA) was extracted and quantified as in Silveira et al. (2004). Briefly, 1 g of powdered tissue was homogenized in a buffer containing 80% (v/v) ethanol, 1% (w/v) polyvinylpyrrolidone-40, and [3H]IAA, used as an internal standard. Samples were incubated and subsequently centrifuged. The supernatant was collected and freeze-dried. Volume was adjusted to 3 mL with water, and the pH was adjusted to 2.5. The organic fraction, obtained following double extraction with ethyl ether, was completely vacuum-dried, redissolved in 150 µL methanol, and filtered through a 0.45-µm membrane. Auxin levels were determined by HPLC in a 5 µm C18 column (Shimadzu Shin-pack CLC ODS), with a fluorescence detector (excitation at 280 nm, emission at 350 nm). Fractions containing IAA were collected and analyzed in the scintillation counter (Packard Tri-Carb) to estimate losses during the procedure.
Leaf Gas-Exchange and Fluorescence Measurements
Gas-exchange and chlorophyll fluorescence parameters were measured in the third leaf completely expanded from 90-d-old plants, as described in Lira et al. (2017), using a portable open gas-exchange system (LI-6400XT system; LI-COR) equipped with an integrated modulated chlorophyll fluorometer (LI-6400-40; LI-COR). Photosynthesis parameters were calculated as in Maxwell and Johnson (2000).
Starch and Soluble Sugar Quantification
Starch and soluble sugars were extracted and determined as described in Bianchetti et al. (2017, 2018) respectively.
Statistical Analyses
Statistical analyses were performed using the Rstudio (https://www.rstudio.com/) and Infostat software (Di Rienzo, 2009). The appropriate test and number of biological replicates used in each experiment are indicated in figure and table descriptions.
Accession Numbers
Sequence data from this article can be found in the Solgenomics database (solgenomics.net) under accession numbers listed in Supplemental Table S5.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Off-vine ripening is affected in SlPIF4-silenced fruits.
Supplemental Figure S2. SlPIF4 silencing affects the number of flowers per truss, but not flowering time.
Supplemental Table S1. Carotenoid content in fruits.
Supplemental Table S2. Yield parameters in wild type and SlPIF4-silenced plants.
Supplemental Table S3. Photosynthesis parameters.
Supplemental Table S4. Soluble sugars.
Supplemental Table S5. Primers used in this work.
ACKNOWLEDGMENTS
We thank Aline Bertinatto Cruz, Amanda Ferreira Macedo, Eglee Igarashi, Viviane Costa, and Silvia Blanco for technical support.
Footnotes
This work was supported by the São Paulo Research Foundation (FAPESP; fellowships 2015/14658-3 (to D.R.), 2016/16764-8 (to B.T.), 2016/02033-1 (to R.Z.), 2017/24354-7 (to R.B.), and 2016/04924-0 (to F.R.R.A.); grant no. 2016/01128-9), the National Council of Scientific and Technological Development (CNPq), the Higher Education Personnel Improvement Coordination (CAPES, Finance Code 001), and the University of São Paulo.
References
- Alba R, Cordonnier-Pratt M-M, Pratt LH (2000) Fruit-localized phytochromes regulate lycopene accumulation independently of ethylene production in tomato. Plant Physiol 123: 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari R, Reuveni M, Evenor D, Nahon S, Shlomo H, Chen L, Levin I (2010) Overexpression of UV-DAMAGED DNA BINDING PROTEIN 1 links plant development and phytonutrient accumulation in high pigment-1 tomato. J Exp Bot 61: 3627–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchetti RE, Cruz AB, Oliveira BS, Demarco D, Purgatto E, Peres LEP, Rossi M, Freschi L (2017) Phytochromobilin deficiency impairs sugar metabolism through the regulation of cytokinin and auxin signaling in tomato fruits. Sci Rep 7: 7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchetti RE, Silvestre Lira B, Santos Monteiro S, Demarco D, Purgatto E, Rothan C, Rossi M, Freschi L (2018) Fruit-localized phytochromes regulate plastid biogenesis, starch synthesis, and carotenoid metabolism in tomato. J Exp Bot 69: 3573–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MT, Maloof JN, Weinig C (2010) Genes underlying quantitative variation in ecologically important traits: PIF4 (PHYTOCHROME INTERACTING FACTOR 4) is associated with variation in internode length, flowering time, and fruit set in Arabidopsis thaliana. Mol Ecol 19: 1187–1199 [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. (2009) The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622 [DOI] [PubMed] [Google Scholar]
- Campos ML, Carvalho RF, Benedito VA, Peres LE (2010) Small and remarkable: The Micro-Tom model system as a tool to discover novel hormonal functions and interactions. Plant Signal Behav 5: 267–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S (2016) Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front Plant Sci 6: 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Yan F, Xu D, Ai K, Yu J, Bao E, Zou Z (2018) Phytochrome B1-dependent control of SP5G transcription is the basis of the night break and red to far-red light ratio effects in tomato flowering. BMC Plant Biol 18: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RF, Campos ML, Pino LE, Crestana SL, Zsögön A, Lima JE, Benedito VA, Peres LE (2011) Convergence of developmental mutants into a single tomato model system: ‘Micro-Tom’ as an effective toolkit for plant development research. Plant Methods 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E (2007) Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Cho LH, Yoon J, An G (2017) The control of flowering time by environmental factors. Plant J 90: 708–719 [DOI] [PubMed] [Google Scholar]
- Cruz AB, Bianchetti RE, Alves FRR, Purgatto E, Peres LEP, Rossi M, Freschi L (2018) Light, ethylene and auxin signaling interaction regulates carotenoid biosynthesis during tomato fruit ripening. Front Plant Sci 9: 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Mustilli AC, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al. (2004) Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing. Plant J 40: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Di Rienzo JA. (2009) Statistical Software for the Analysis of Experiments of Functional Genomics. RDNDA, Argentina [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández V, Takahashi Y, Le Gourrierec J, Coupland G (2016) Photoperiodic and thermosensory pathways interact through CONSTANS to promote flowering at high temperature under short days. Plant J 86: 426–440 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu Y-S, Fernie AR, Zamir D (2004) Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Nakano T, Ito Y (2011) Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Collani S, Horrer D, Schmid M (2015) Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J 84: 949–962 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Kumar SV (2017) DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Reports 18: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramegna G, Rosado D, Sánchez Carranza AP, Cruz AB, Simon-Moya M, Llorente B, Rodríguez-Concepcíon M, Freschi L, Rossi M (2019) PHYTOCHROME-INTERACTING FACTOR 3 mediates light-dependent induction of tocopherol biosynthesis during tomato fruit ripening. Plant Cell Environ 42: 1328–1339 [DOI] [PubMed] [Google Scholar]
- Gupta SK, Sharma S, Santisree P, Kilambi HV, Appenroth K, Sreelakshmi Y, Sharma R (2014) Complex and shifting interactions of phytochromes regulate fruit development in tomato. Plant Cell Environ 37: 1688–1702 [DOI] [PubMed] [Google Scholar]
- Jung J-H, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development In Timmermans MCP, ed, Current Topics in Developmental Biology, Vol Vol 91 Elsevier, Amsterdam, pp 29–66 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Krieger U, Lippman ZB, Zamir D (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42: 459–463 [DOI] [PubMed] [Google Scholar]
- Kudo M, Kidokoro S, Yoshida T, Mizoi J, Todaka D, Fernie AR, Shinozaki K, Yamaguchi-Shinozaki K (2017) Double overexpression of DREB and PIF transcription factors improves drought stress tolerance and cell elongation in transgenic plants. Plant Biotechnol J 15: 458–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol 52: 1315–1329 [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CCR, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E (2014) PIFs: Systems integrators in plant development. Plant Cell 26: 56–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JLS, Gee CW, Sairanen I, Ljung K, Nemhauser JL (2012) An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol 160: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira BS, Gramegna G, Trench BA, Alves FRR, Silva EM, Silva GFF, Thirumalaikumar VP, Lupi ACD, Demarco D, Purgatto E, et al. (2017) Manipulation of a senescence-associated gene improves fleshy fruit yield. Plant Physiol 175: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira BS, Rosado D, Almeida J, de Souza AP, Buckeridge MS, Purgatto E, Guyer L, Hörtensteiner S, Freschi L, Rossi M (2016) Pheophytinase knockdown impacts carbon metabolism and nutraceutical content under normal growth conditions in tomato. Plant Cell Physiol 57: 642–653 [DOI] [PubMed] [Google Scholar]
- Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101: 9897–9902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, D’Andrea L, Ruiz-Sola MA, Botterweg E, Pulido P, Andilla J, Loza-Alvarez P, Rodriguez-Concepcion M (2016) Tomato fruit carotenoid biosynthesis is adjusted to actual ripening progression by a light-dependent mechanism. Plant J 85: 107–119 [DOI] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—A practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- McDonald MS. (2003) Photobiology of Higher Plants. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Molinero-Rosales N, Latorre A, Jamilena M, Lozano R (2004) SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218: 427–434 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière J-M, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the Evening Complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E (2018) Phytochromes and phytochrome interacting factors. Plant Physiol 176: 1025–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino LE, Lombardi-Crestana S, Azevedo MS, Scotton DC, Borgo L, Quecini V, Figueira A, Peres LE (2010) The Rg1 allele as a valuable tool for genetic transformation of the tomato ‘Micro-Tom’ model system. Plant Methods 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Almeida J, Otaiza SN, Duffy T, da Silva JVC, de Godoy F, Asís R, Bermúdez L, Fernie AR, Carrari F, et al. (2013) Transcriptional regulation of tocopherol biosyntheis in tomato. Plant Mol Biol 81: 309–325 [DOI] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M (2016) Molecular and genetic control of plant thermomorphogenesis. Nat Plants 2: 15190. [DOI] [PubMed] [Google Scholar]
- Rodriguéz GR, Moyseenko JB, Robbins MD, Morejón NH, Francis DM, van der Knaap E (2010) Tomato Analyzer: A useful software application to collect accurate and detailed morphological and colorimetric data from two-dimensional objects. J Vis Exp 37: e1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado D, Gramegna G, Cruz A, Lira BS, Freschi L, de Setta N, Rossi M (2016) Phytochrome Interacting Factors (PIFs) in Solanum lycopersicum: Diversity, evolutionary history and expression profiling during different developmental processes. PLoS One 11: e0165929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF (2009) Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Jeong J, Kang M-Y, Kim J, Paek N-C, Choi G (2014) Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun 5: 4636. [DOI] [PubMed] [Google Scholar]
- Seaton DD, Smith RW, Song YH, MacGregor DR, Stewart K, Steel G, Foreman J, Penfield S, Imaizumi T, Millar AJ, et al. (2015) Linked circadian outputs control elongation growth and flowering in response to photoperiod and temperature. Mol Syst Biol 11: 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Zhang H, Song X, Jiang Y, Liang R, Li G (2018) Functional characterization of the maize Phytochrome-interacting factors PIF4 and PIF5. Front Plant Sci 8: 2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva GFF, Silva EM, Correa JPO, Vicente MH, Jiang N, Notini MM, Junior AC, De Jesus FA, Castilho P, Carrera E, et al. (2019) Tomato floral induction and flower development are orchestrated by the interplay between gibberellin and two unrelated microRNA-controlled modules. New Phytol 221: 1328–1344 [DOI] [PubMed] [Google Scholar]
- Silveira V, Balbuena TS, Santa-Catarina C, Floh EI, Guerra MP, Handro W (2004) Biochemical changes during seed development in Pinus taeda L. Plant Growth Regul 44: 147–156 [Google Scholar]
- Song Y, Yang C, Gao S, Zhang W, Li L, Kuai B (2014) Age-triggered and dark-induced leaf senescence require the bHLH transcription factors PIF3, 4, and 5. Mol Plant 7: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB (2017) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162–168 [DOI] [PubMed] [Google Scholar]
- Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, Roustan J-P, Bouzayen M, Giuliano G, Chervin C (2015) Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol 15: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines BC, Youn Y, Duarte MI, Harmon FG (2014) The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J Exp Bot 65: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, et al. (2012) Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci USA 109: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW (2004) Phytochrome signaling mechanism. The Arabidopsis Book 3: 3, doi:10.1186-4811-3-12.ris [DOI] [PMC free article] [PubMed]
- Wang L, Wu LM, Greaves IK, Zhu A, Dennis ES, Peacock WJ (2017) PIF4-controlled auxin pathway contributes to hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: E3555–E3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu Y, Wang H, Ma X, Wang B, Wu G, Wang H (2017) Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat Commun 8: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P-B, Lian H-L, Wang W-X, Xu F, Yang H-Q (2016) Pivotal roles of the phytochrome-interacting factors in cryptochrome signaling. Mol Plant 9: 496–497 [DOI] [PubMed] [Google Scholar]
- Yan J, Liu Y, Wang K, Li D, Hu Q, Zhang W (2018) Overexpression of OsPIL1 enhanced biomass yield and saccharification efficiency in switchgrass. Plant Sci 276: 143–151 [DOI] [PubMed] [Google Scholar]
- Zhang S, Jiao Z, Liu L, Wang K, Zhong D, Li S, Zhao T, Xu X, Cui X (2018) Enhancer-promoter interaction of SELF PRUNING 5G shapes photoperiod adaptation. Plant Physiol 178: 1631–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Z, Chen Y, He J-X, Bi Y (2015) PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci 237: 57–68 [DOI] [PubMed] [Google Scholar]