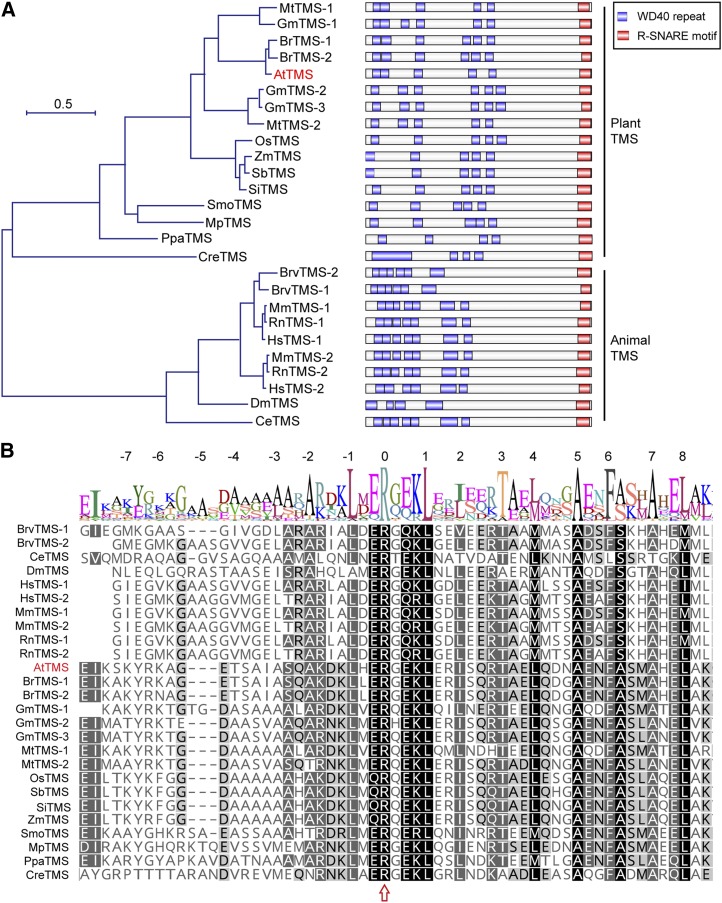
Figure 1.
AtTMS is homologous to animal TMSs. A, Left, Phylogenetic relationship among AtTMS, other plant TMS homologs, and animal TMS proteins. The phylogenic tree was constructed using MEGA version 7.0. Species abbreviations are as follows: Mt, Medicago truncatula; Gm, Glycine max; Br, Brassica rapa; At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays; Sb, Sorghum bicolor; Si, Setaria italica; Smo, Selaginella moellendorfii; Mp, Marchantia polymorpha; Ppa, Physcomitrella patens; Cre, Chlamydomonas reinhardtii; Mm, Mus musculus; Rn, Rattus norvegicus; Hs, Homo sapiens; Brv, Brachydanio rerio var; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans. The sequence alignment used for this analysis is available as Supplemental Data Set S1. Right, The domain structure of each TMS family member is presented next to the corresponding protein. B, Sequence alignment of R-SNARE motif of TMS proteins. Numbers at the top indicate helical layers formed during SNARE complex assembly. The arrow indicates the Arg residue (R) at layer 0.