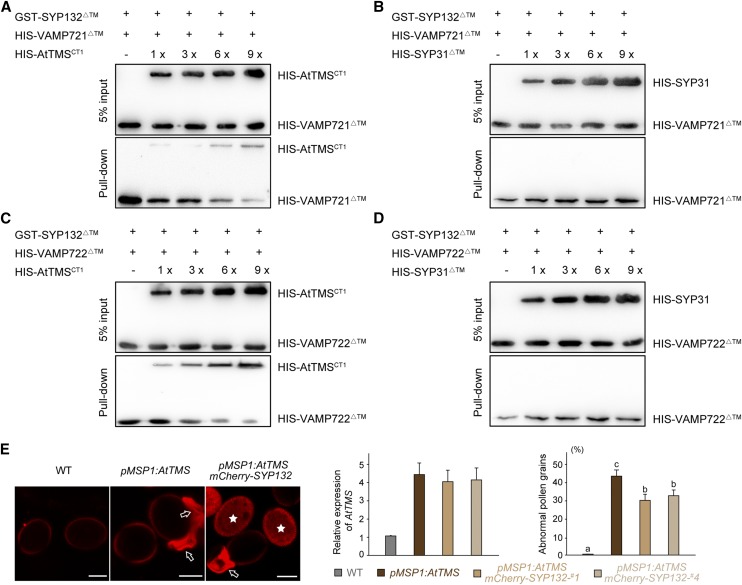
Figure 8.
AtTMS competes with VAMP721/722 for the binding to SYP132, SYP132 partially rescues defects of pMSP1:AtTMS pollen. A and B, SYP132 partially rescues defects of pMSP1:AtTMS pollen (A), and AtTMSCT1 competes with VAMP721ΔTM for binding to SYP132ΔTM (B). In each lane, 6 µg of GST-SYP132ΔTM proteins coupled to beads was incubated with 2 µg of VAMP721ΔTM and increasing amounts (1, 3, 6, and 9 µg) of HIS-AtTMSCT1 (A) or 2 µg of VAMP721ΔTM with increasing amounts of Golgi resident Qa-SNARE HIS-SYP31ΔTM (B). After washing, bound proteins were detected using anti-His antibodies. C and D The same method used in A and B was adopted. Similarly, AtTMSCT1 was shown to compete with VAMP722ΔTM for the binding to SYP132ΔTM. E, Confocal images of pollen grains from the wild type (WT), pMSP1:AtTMS, and pMSP1:AtTMS + pSYP132:mCherry-SYP132. Arrows and asterisks indicate collapsed pollen and mCherry-SYP132-expressing pollen, respectively. Note that collapsed pollen grains display specific red fluorescence. In the left-hand graph, mRNA used for RT-qPCR was extracted from flower buds, and AtTMS expression relative to that of the wild type is presented. In the right-hand graph, one pMSP1::AtTMS line showed approximately 45% abnormal pollen grains, whereas two lines of pSYP132::mCherry-SYP132 + pMSP1::AtTMS showed about 30% abnormal pollen grains. n > 200 pollen was counted for each line. Values represent means ± sd of three independent experiments. Statistical significance using Duncan’s ANOVA is indicated by lowercase letters (P < 0.05). Bars = 10 μm.