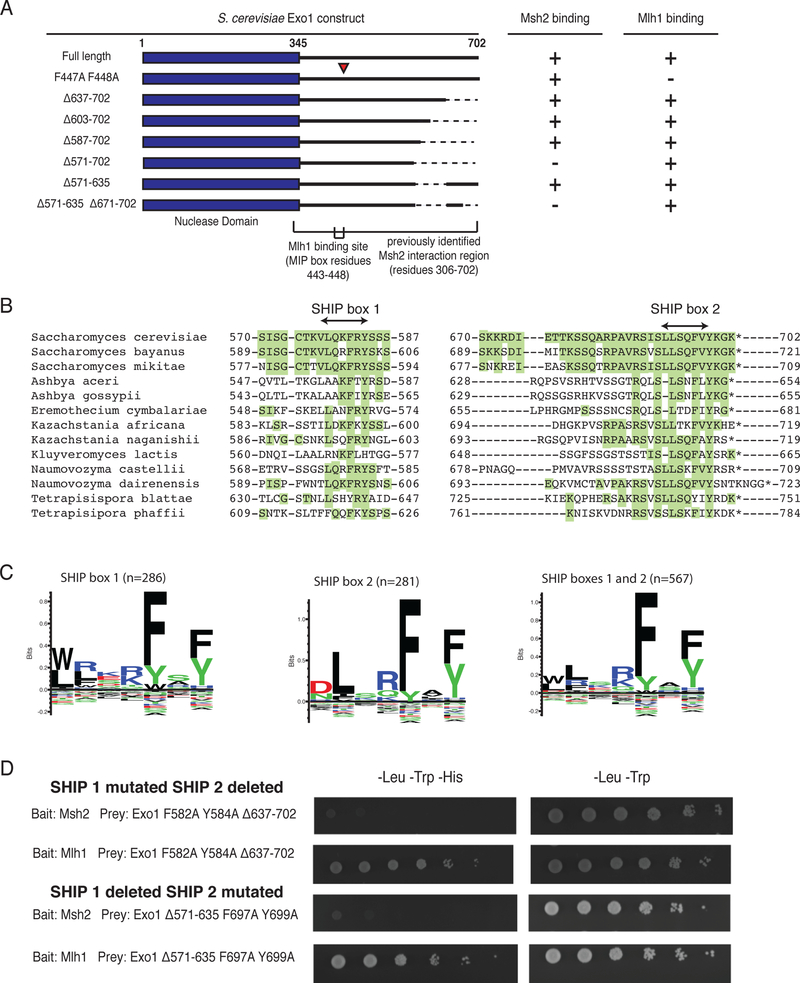
Figure 1. Two regions in the S. cerevisiae Exo1 C-terminal tail mediate Msh2 interaction.
A. Summary of yeast two-hybrid interactions with various Exo1 deletion constructs in prey vectors and their interactions with bait vectors encoding Msh2 or Mlh1. The Exo1-F447A,F448A variant disrupts the Exo1 MIP box and the Mlh1 binding interaction. The Exo1Δ571–702 and Exo1Δ571–635,Δ671–702 disrupt the Msh2 binding sites, but not the Exo1-Mlh1 interaction. All experiments were independently repeated a minimum of 4 times. B. Sequence alignment of the Msh2-binding regions of S. cerevisiae Exo1 with closely related fungal species in the Saccharomycotina are displayed so that residues identical to S. cerevisiae are highlighted in green. C. Sequence logos generated by Seq2Logo (ref) for alignments of SHIP box 1, SHIP box 2, and both SHIP boxes based on an alignment of 291 fungal species; note that the number of sequences for each SHIP box is lower than the total number of fungal species analysed due to gene annotation errors and/or lack of conservation of specific SHIP boxes. D. Yeast two-hybrid analysis reveals that the Exo1-F582A,Y584A,Δ637–702 and Exo1Δ571–635,F697A,Y699A constructs failed to interact with the Msh2 bait (lack of growth on –Leu –Trp –His medium as compared to growth on the control –Leu –Trp medium) but still retained interaction with the Mlh1 bait. All experiments were independently repeated a minimum of 4 times.