Abstract
Sample multiplexing facilitates scRNA-seq by reducing costs and artifacts such as cell doublets. However, universal and scalable sample barcoding strategies have not been described. We therefore developed MULTI-seq: multiplexing using lipid-tagged indices for single-cell and single-nucleus RNA sequencing. MULTI-seq reagents can barcode any cell type or nucleus from any species with an accessible plasma membrane. The method involves minimal sample processing, thereby preserving cell viability and endogenous gene expression patterns. MULTI-seq enables doublet identification, which improves data quality and increases cell throughput by minimizing the negative consequences of Poisson droplet loading. MULTI-seq sample classifications additionally identify cells with low RNA content that would otherwise be discarded by standard quality-control workflows. We use MULTI-seq to track the dynamics of T-cell activation, perform a 96-plex perturbation experiment with primary human mammary epithelial cells, and multiplex cryopreserved tumors and metastatic sites isolated from a patient-derived xenograft mouse model of triple-negative breast cancer.
INTRODUCTION
Single-cell and single-nucleus RNA sequencing (scRNA-seq, snRNA-seq) have emerged as powerful technologies for interrogating the heterogeneous transcriptional profiles of multicellular systems. Early scRNA-seq workflows were limited to analyzing tens to hundreds of single-cell transcriptomes at a time1,2. With the advent of single-cell sequencing technologies based on microwell3, split-pool barcoding4,5, and droplet-microfluidics6–9 the parallel transcriptional analysis of 103–105 cells or nuclei is now routine. This increase in cell-throughput has catalyzed efforts to characterize the composition of whole organs10 and entire organisms4,11.
These technologies will increasingly be used to reveal the mechanisms by which cell populations interact to promote development, homeostasis, and disease. This shift from descriptive to mechanistic analyses requires integrating spatiotemporal information, diverse perturbations, and experimental replicates in order to draw strong conclusions12,13. While existing methods can assay many thousands of cells, sample-specific barcodes (e.g., Illumina library indices) are incorporated at the very end of standard library preparation workflows, which limits scRNA-seq sample-throughput due to reagent costs and the physical constraints of droplet microfluidics devices. Sample multiplexing approaches address this limitation by labeling cells with sample-specific barcodes prior to pooling and single-cell isolation. Several multiplexing methods have been described that distinguish samples using pre-existing genetic diversity14, or introduce sample barcodes using either genetic15–20 or non-genetic21–23 mechanisms. However, each of these methods has liabilities, including issues with scalability, universality, and the potential to introduce secondary perturbations to experiments.
We identified lipid- and cholesterol-modified oligonucleotides (LMOs, CMOs) as reagents that circumvent many of the limitations of other sample multiplexing techniques. We previously described LMO and CMO scaffolds that rapidly and stably incorporate into the plasma membrane of live cells by step-wise assembly24. Here, we adapt LMOs and CMOs into MULTI-seq – scRNA-seq and snRNA-seq sample multiplexing using lipid-tagged indices. MULTI-seq localizes sample barcodes to live cells and nuclei regardless of species or genetic background. MULTI-seq is non-perturbative, rapid, and involves minimal sample processing. Here, MULTI-seq simplicity and modularity enabled the analysis of a T-cell activation time-course, 96 human mammary epithelial cell (HMEC) culture conditions, and cryopreserved primary cells isolated from patient-derived xenograft (PDX) mouse models at varying stages of metastatic progression.
RESULTS
MULTI-seq overview:
MULTI-seq localizes DNA barcodes to plasma membranes by hybridization to an ‘anchor’ LMO. The ‘anchor’ LMO associates with membranes through a hydrophobic 5’ lignoceric acid amide. Subsequent hybridization to a ‘co-anchor’ LMO incorporating a 3’ palmitic acid amide increases the hydrophobicity of the complex and thereby prolongs membrane retention (Fig. 1A). MULTI-seq sample barcodes include a 3’ poly-A capture sequence, an 8 bp sample barcode, and a 5’ PCR handle necessary for library preparation and anchor hybridization. Cells or nuclei carry membrane-associated MULTI-seq barcodes into emulsion droplets where the 3’ poly-A domain mimics endogenous transcripts during hybridization to mRNA capture beads. Endogenous transcripts and MULTI-seq barcodes are then linked to a common cell- or nucleus-specific barcode during reverse transcription, which enables sample demultiplexing. MULTI-seq barcode and endogenous expression libraries are separated by size selection prior to next-generation sequencing library construction, enabling pooled sequencing at user-defined proportions (Experimental Methods). The same strategy can be applied to commercially-available CMOs.
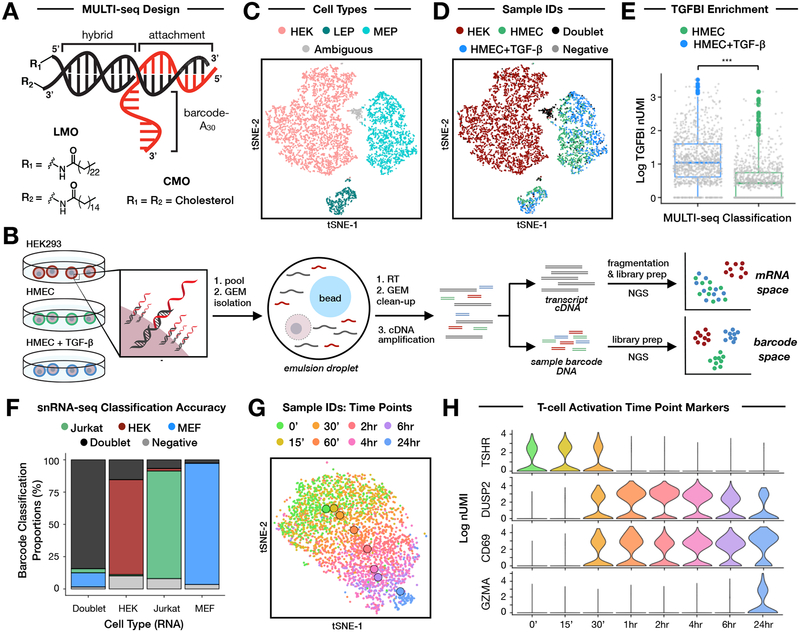
Figure 1: MULTI-seq demultiplexes cell types, culture conditions, and time points for single-cell and single-nucleus RNA sequencing.
(A) Diagram of the anchor/co-anchor LMO and CMO scaffolds (black) with hybridized sample barcode oligonucleotide (red). LMOs and CMOs are distinguished by their unique lipophilic moieties (e.g., lignoceric acid, palmitic acid, or cholesterol).
(B) Schematic overview of a proof-of-concept single-cell RNA sequencing experiment using MULTI-seq. Three samples (HEKs and HMECs with and without TGF-β stimulation) were barcoded with either LMOs or CMOs and sequenced alongside unlabeled controls. Cells were pooled together prior to scRNA-seq. Next-generation sequencing produces two UMI count matrices corresponding to gene expression and barcode abundances.
(C) Cell type annotations for LMO-labeled cells demonstrate separation between HEKs (pink), MEPs (cyan), and LEPs (dark teal) in gene expression space (see Fig. S2A). Ambiguous cells positive for multiple marker genes are displayed in grey. n = 6,186 MULTI-seq barcoded cells.
(D) MULTI-seq sample classifications for HEKs (dark red), unstimulated HMECs (green), and TGF-β-stimulated HMECs (blue) match cell state annotations. Cells classified as doublets (black) predominantly overlap with ambiguously-annotated cells. n = 6,186 MULTI-seq barcoded cells.
(E) TGF-β-stimulated HMECs (blue) exhibited elevated TGFBI expression relative to unstimulated HMECs (green). *** = Wilcoxon rank sum test (two-sided), p <= 10−16. n = 1,950 MULTI-seq barcoded HMECs. Data are represented as mean ± SEM.
(F) Single nucleus MULTI-seq sample classification proportions for each cell type identified by clustering in gene expression space (see Fig. S2E–G). n = 5,894 MULTI-seq barcoded nuclei.
(G) MULTI-seq sample classifications illuminate temporal gene expression patterns in Jurkat cells following activation with ionomycin and PMA for varying amounts of time. Time-point centroids in gene expression space are denoted with larger circles. n = 3,709 Jurkat nuclei.
(H) Violin plots of gene expression marking different stages of Jurkat cell activation. n = 3,709 Jurkat nuclei.
We used flow cytometry to evaluate whether LMOs and CMOs predictably label and minimally exchange between live cells at typical sample preparation temperatures of 4 °C (Fig. S1A, S1B). Identical experiments were also performed using freshly-isolated nuclei (Fig. S1C, S1D). These data revealed that LMOs exhibit longer membrane residency times than CMOs on live-cell membranes at 4 °C, whereas LMOs and CMOs exchange comparably between live cells at room temperature, suggesting cells should be maintained on ice to achieve optimal sample multiplexing results (Fig. S1E). For nuclei, both oligonucleotide conjugates showed minimal exchange between nuclear membranes (Fig. S1D), however, bovine serum albumin (BSA) in nuclei isolation buffer specifically quenched LMOs, reducing labeling efficiency (Fig. S1B). While problematic during nuclei labeling, we reasoned that LMO quenching could be strategically employed during live cell labeling to reduce off-target barcoding and potentially minimize washes prior to sample pooling. Indeed, we found that diluting LMO-labeling reactions with 1% BSA in PBS resulted in minimal off-target labeling following pooling (< 1% of primary labeling signal), which was 18-fold lower than dilution with PBS (Fig. S1F).
MULTI-seq enables scRNA-seq sample demultiplexing:
We tested the capacity of MULTI-seq to demultiplex scRNA-seq samples by performing a proof-of-concept experiment using HEK293 cells (HEKs) and primary human mammary epithelial cells (HMECs) cultured in the presence or absence of TGF-β (Fig. 1B). Cells were trypsinized, barcoded with LMOs or CMOs, and pooled prior to droplet microfluidic-emulsion with the 10X Genomics Chromium system. In parallel, we prepared un-barcoded replicates to test whether MULTI-seq influenced gene expression or mRNA capture efficiency.
Following data pre-processing (Computational Methods), we analyzed a final scRNA-seq dataset containing 14,377 total cells. We identified clusters in gene expression space according to known markers for HEKs as well as the two cellular components of HMECs, myoepithelial (MEPs) and luminal epithelial cells (LEPs, Fig. 1C, Fig. S2A). Projecting MULTI-seq barcode classifications onto gene expression space for LMO-labeled (Fig. 1D) and CMO-labeled cells (Fig. S2B) illustrates that both membrane scaffolds successfully demultiplexed each sample. HMECs predicted to have been cultured with TGF-β exhibited enriched TGFBI expression (Fig. 1E). Importantly, RNA and MULTI-seq barcode UMI counts were not negatively correlated, demonstrating that MULTI-seq does not impair mRNA capture (Fig. S2C). However, we observed transcriptional changes in CMO-labeled HEKs (Fig. S2D, Table S1) that were absent in LMO-labeled HEKs.
Demultiplexing single nucleus RNA-seq (snRNA-seq) and time-course experiments:
snRNA-seq is widely used for the analyses of solid tissues that are difficult to dissociate25. We explored whether MULTI-seq could demultiplex snRNA-seq samples by purifying nuclei from HEKs and mouse embryonic fibroblasts (MEFs) and labeling each pool of nuclei with LMOs or CMOs prior to snRNA-seq. In parallel, we multiplexed Jurkat cells treated with ionomycin and phorbol 12-myristate 13-acetate (PMA) at eight time points (0–24 hours) to track T-cell activation dynamics (Fig. S2E). MULTI-seq sample classifications matched their intended cell type clusters in gene expression space (Fig. S2F, Fig. S2G) with a ~0.5% misclassification rate (Fig. 1F). Notably, MULTI-seq classifications were species-specific and predicted ~85% of mouse-human doublets, which approximates the theoretical doublet detection limit of ~92%. Matching live-cell results, MULTI-seq barcoding did not impair mRNA capture (Fig. S2H). In contrast to live-cell results, both CMO- and LMO-labeled nuclei were transcriptionally indistinguishable from unbarcoded controls (Fig. S2I). Moreover, CMO-labeled nuclei had higher average signal-to-noise ratio (SNR) and total number of barcode UMIs relative to LMO-labeled nuclei (Table S2), consistent with previous flow cytometry results.
Upon demultiplexing individual time points along the trajectory of T-cell activation (Fig. 1G), we observed multiple literature-supported transcriptional dynamics (Fig. 1H). For example, genes undergoing early down-regulation (e.g., TSHR26) and transient (e.g., DUSP227), sustained (e.g., CD6928), and late (e.g., GRZA29) up-regulation were readily identified in the data.
MULTI-seq identifies doublets in scRNA-seq data:
We next sought to demonstrate MULTI-seq scalability by multiplexing 96 unique HMEC samples spanning a range of microenvironmental conditions. We exposed duplicate cultures consisting of MEPs, LEPs, and both cell types grown in M87A media30 without EGF to 15 physiologically-relevant signaling molecules31 or signaling molecule combinations (Fig. S3A). We barcoded each sample before pooling and loaded cells across three 10X microfluidics lanes, resulting in a 32-fold reduction in reagent use relative to standard practices.
To classify HMECs into sample groups, we implemented a sample classification workflow inspired by previous strategies15,16,21 (Computational Methods, Supplemental Materials, Fig. S3) which identified 76 sample groups consisting of 26,439 total cells (Fig. S4). Each group was exclusively enriched for a single barcode (Fig. 2A, left, Fig. S3C) an average of ~199-fold above the most abundant off-target barcode (Fig. S3D). Unlike sample multiplexing data with relatively few samples, MULTI-seq-defined doublets localized to the peripheries of singlet clusters in barcode space for this experiment (Fig. 2A, right). We suspected missing barcodes resulted from handling errors (Fig. S3B, Supplemental Materials), as a technical replicate yielded all 96 sample groups (Fig. S3E–G).
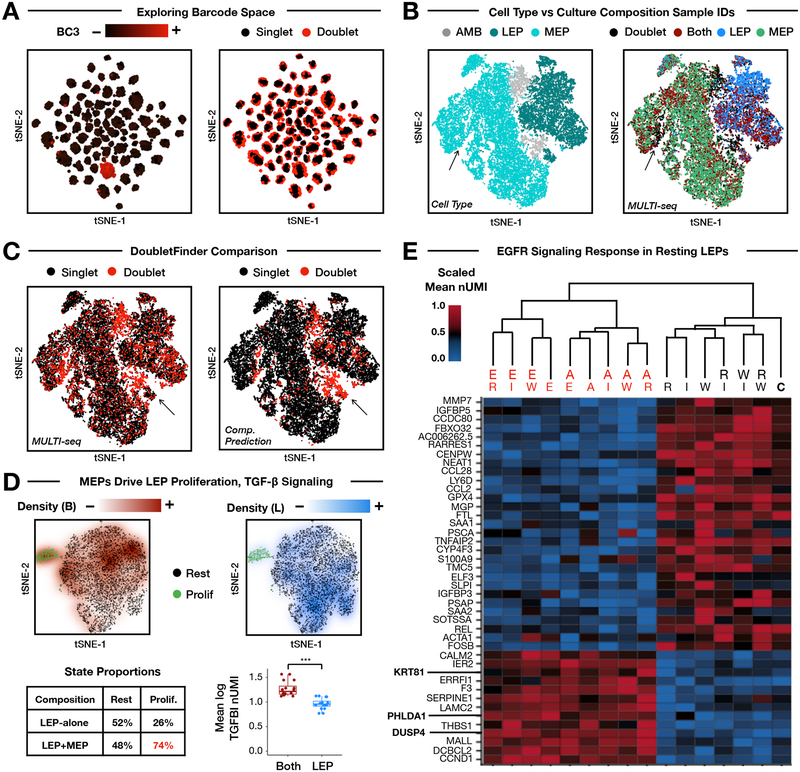
Figure 2: MULTI-seq barcoding of multiplexed HMEC culture conditions.
(A) Barcode UMI abundances (left) and doublet classifications (right) mapped onto barcode space. MULTI-seq barcode #3 is used as a representative example. Doublets localize to the peripheries of sample groups in large-scale sample multiplexing experiments. n = 25,166 cells.
(B) Cell state annotations demonstrate separation between MEPs (cyan) and LEPs (dark teal) in gene expression space (left, see Fig. S5A). Ambiguous cells positive for multiple marker genes are displayed in grey. MULTI-seq classifications grouped by culture composition (right) — e.g., LEP-alone (blue), MEP-alone (green), and both cell types together (dark red) — match cell state annotations. Discordant region where annotated MEPs are classified as doublets by MULTI-seq is indicated with arrows. n = 25,166 cells.
(C) MULTI-seq doublet classifications (left) and computational predictions produced by DoubletFinder (right) largely overlap in gene expression space. Discordant region where DoubletFinder-defined doublets that are classified as singlets by MULTI-seq indicated with arrows. n = 25,166 cells.
(D) MEP co-culture induces LEP proliferation and TGF-β signaling. Clusters corresponding to resting (black) and proliferative (green) LEPs are identifiable in gene expression space (Fig. S5B). Projecting sample classification densities onto gene expression space for co-cultured LEPs (dark red, top left) and LEPs cultured alone (blue, top right) illustrates that co-cultured LEPs are enriched in the proliferative state (table, bottom left). Co-cultured LEPs also express more TGFBI than LEPs cultured alone. Each point represents an average of LEPs grouped according growth factor condition. *** = Wilcoxon rank sum test (two-sided), p = 3.1×10−6. n = 32 signaling molecule condition groups. Data are represented as mean ± SEM.
(E) Hierarchical clustering and heat map analysis of resting LEPs grouped by treatment. Emphasized genes are known EGFR signaling targets. RNA UMI abundances are scaled from 0–1 for each gene. Values correspond to the average expression within each signaling molecule treatment group. Dendrogram labels: E = EGF, W = WNT4, A = AREG, I = IGF-1, R = RANKL, C = Control.
To assess demultiplexing accuracy, we grouped MULTI-seq classifications according to cell type composition (e.g., MEPs alone, LEPs alone, or both) and visualized these groups in gene expression space. Unsupervised clustering and marker analysis of the resulting transcriptome data distinguished LEPs from MEPs along with a subset of ambiguous cells expressing markers for both cell types (Fig. 2B, left, Fig. S5A). MULTI-seq classifications matched their expected cell type clusters (Fig. 2B, right), while cells co-expressing MEP and LEP markers were predominantly defined as doublets. MULTI-seq identified doublets that were overlooked when predicting doublets using marker genes (Fig. 2B, arrow). Additionally, MULTI-seq doublet classifications generally agreed with computational predictions (Fig. 2C, Sensitivity = 0.283 Specificity = 0.965), with the exception of ‘homotypic’ doublets – i.e., doublets formed from transcriptionally-similar cells – to which computational doublet detection techniques are insensitive32,33 (Supplemental Materials). Moreover, DoubletFinder erroneously classified proliferative LEPs as doublets (Fig. 2C, arrow), illustrating how computational doublet inference performance suffers when applied to datasets with low cell type numbers32,33.
MULTI-seq identifies transcriptional responses to co-culture conditions and signaling molecules:
Sample demultiplexing, doublet removal, and quality-control filtering resulted in a final scRNA-seq dataset including 21,753 total cells, revealing two transcriptional responses linked to culture composition. First, we observed that LEPs co-cultured with MEPs exhibited enriched proliferation relative to LEPs cultured alone (Fig. 2D, Fig. S5B). In contrast, MEPs were equally proliferative when cultured alone or with LEPs (Fig. S5C). Second, we observed that non-proliferative co-cultured MEPs and LEPs are enriched for TGFBI expression relative to MEPs and LEPs cultured alone (Fig. 2D, bottom right, Fig. S5D).
We next used hierarchical clustering to assess how LEPs or MEPs responded to signaling molecule exposure. HMECs exposed to the EGFR ligands AREG and EGF exhibited gene expression profiles that were significantly different from control cells. AREG- and EGF-stimulated LEPs expressed increased levels of EGFR signaling genes (e.g., DUSP434) and genes up-regulated in HER2+ breast cancers (e.g., PHLDA135, Fig. 2E) relative to control LEPs. AREG- and EGF-stimulated MEPs also express high levels of known EGFR-regulated genes (e.g., ANGPTL436, Fig. S5E).
MULTI-seq identifies low-RNA cells in cryopreserved, primary PDX samples:
Using scRNA-seq to analyze archival primary tissue samples is often difficult because these samples can have low cell viability that is compounded during cryopreservation, thawing, enzymatic digestion, and scRNA-seq sample preparation. We investigated whether the rapid and non-perturbative nature of MULTI-seq barcoding would enable cryopreserved tissue multiplexing using samples dissected from a PDX mouse model of metastatic triple-negative breast cancer37. In this model system, the diameter of primary tumors was used as a proxy for metastatic progression in the lung (Fig. S6A). We barcoded 9 distinct samples representing primary tumors and lungs from early- and mid-stage PDX mice (in duplicate), one late-stage PDX mouse, and a single lung from an immunodeficient mouse without tumors (Fig. 3A). We then pooled FACS-enriched populations of barcoded hCD298+ human metastases with mCD45+ mouse immune cells prior to “super-loading” a single 10X Genomics microfluidics lane.
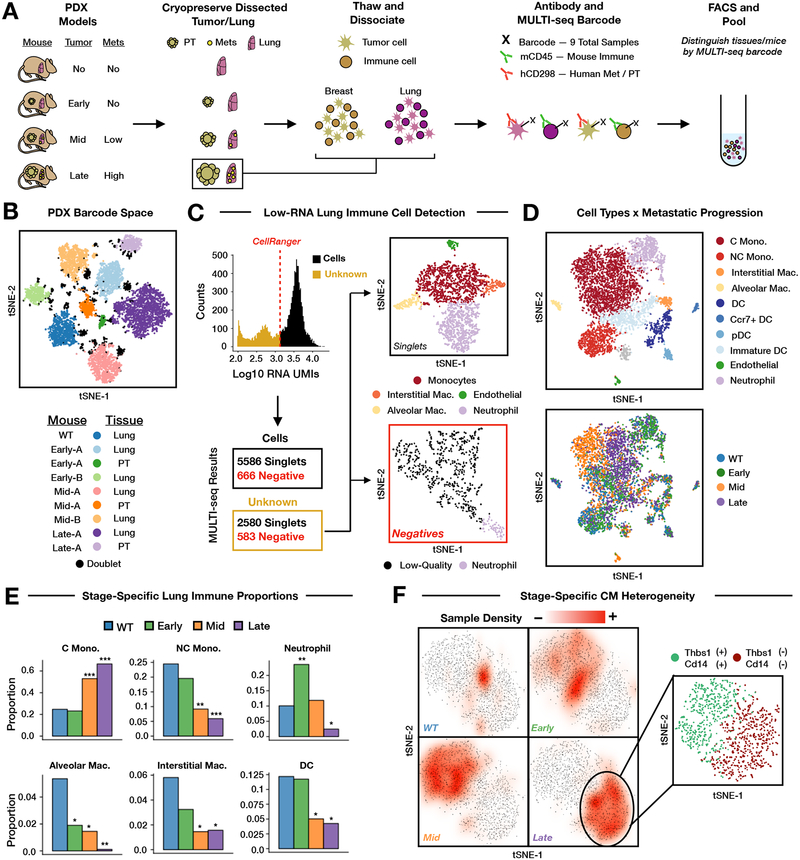
Figure 3: PDX sample multiplexing demonstrates low-RNA cell detection, reveals immune cell proportional shifts and classical monocyte heterogeneity in the progressively metastatic lung.
(A) Schematic overview of PDX experiment.
(B) MULTI-seq sample classifications (WT, early, mid, late tumor progression) mapped onto barcode space. Replicate tissues are denoted as ‘A’ or ‘B’. n = 10,427 cells.
(C) MULTI-seq classifications facilitate low-RNA and low-quality cell deconvolution. CellRanger discards cells barcodes with low RNA UMI counts (red dotted line). Gene expression profiles for classified low-RNA cells reflect established immune cell types (top right, see Fig. S6F). Unclassified low-RNA cells resemble low-quality single-cell transcriptomes (bottom right, see Table S4). n = 2,580 (classified), 583 (unclassified) cells.
(D) Cell state annotations (top) and tumor stages (bottom) for lung immune cells in gene expression space. Mono. = monocyte, C = classical, NC = non-classical, Mac. = macrophage, DC = dendritic cell, pDC = plasmacytoid DC. Cells with undeterminable annotations displayed in grey. n = 5,965 cells.
(E) Statistically-significant shifts in lung immune cell type proportions for each tumor stage relative to WT. Two-proportion z-test with Bonferroni multiple comparisons adjustment, * = 0.05 > p > 10−10; ** = 10−10 > p > 10−20; *** = p < 10−20. n = 44 tumor-stage/cell type groups. Statistically-insignificant proportional shifts omitted.
(F) Subsetted classical monocyte gene expression space overlaid with sample classification densities corresponding to tumor stage. Inset illustrates heterogeneity within late-stage classical monocytes characterized by differential expression of Thbs1 and Cd14. n = 2,496 (all), 1,087 (inset) cells.
Quality-control filtering, sample classification, and doublet removal resulted in a final scRNA-seq dataset of 9,110 mouse and human singlets spanning all 9 samples (Fig. 3B, Fig. S6B). Under the conditions tested, barcode SNR was largely invariant to inter-sample differences in total cell number and viability (Fig. S6C, Table S3). Classification accuracy was supported by tissue-specific gene expression patterns (Fig. S6D) and comparisons to FACS enrichment results (Fig. S6E). Additionally, MULTI-seq classifications identified high-quality single-cell transcriptomes that would have been discarded using standard quality-control workflows (e.g., CellRanger RNA UMI inflection point threshold = 1350, Fig. 3C). When comparing cells with 100–1350 RNA UMIs, classified cells included immune cell types that are difficult to detect using single-cell and bulk transcriptomics (e.g., neutrophils38). Strikingly, 90.8% of sequenced neutrophils would have been discarded by CellRanger. In contrast, unclassified low-RNA cells had poor-quality gene expression profiles predominantly corresponding to broken cells39 (Table S4).
Characterizing the lung immune response to metastatic progression:
We next sought to describe how lung immune cells respond to metastatic progression. Beginning with a dataset comprised of 5,690 mCD45+ cells, we identified gene expression profiles associated with neutrophils, monocytes and macrophages (alveolar, interstitial, and (non)-classical monocytes), dendritic cells (mature, immature, Ccr7+, and plasmocytoid DCs), and endothelial cells10,40 (Fig. 3C, top, Fig. S6F). The use of immunodeficient PDX mice resulted in a lack of lymphocytes (e.g., T, B, and NK cells).
We observed literature-supported changes in immune cell proportions (Fig. 3D) and transcriptional state (Fig. 3E) at each tumor stage. For instance, neutrophils were enriched in early-stage PDX mice while alveolar macrophages were depleted over the course of metastasis41,42. Moreover, stage-specific transcriptional heterogeneity among classical monocytes (CMs, Fig. 3F) reflects previous descriptions of lung CM state transitions in PDX breast cancer models43.
Unsupervised clustering of CMs cleanly resolved cells from each tumor stage (Fig. S7G), enabling the identification of genes upregulated in CMs during metastatic progression (Table S5). Notably, clustering also revealed that CMs from late-stage PDX mice fell into two distinct transcriptional states discernible by Cd14 expression (Fig. 3F, inset, Table S6) matching previous observations44. Genes that are differentially-expressed between CM subsets include genes known to influence metastatic progression43,45,46 (e.g., Thbs1, S100a8/9, and Wfdc21). To discern whether the results were primarily attributable to inter-mouse variability, we used Earth Mover’s Distance (EMD)47 to quantify the magnitude of transcriptional dissimilarity between lung CMs from each mouse and tumor stage. These results illustrate that CMs from early- and mid-stage mouse replicates (scaled EMD = 0.16) were more similar than CMs from distinct tumor stages (scaled EMD = 0.69).
DISCUSSION
MULTI-seq is an ideal sample multiplexing approach because it is scalable, universal, and improves scRNA-seq data quality. MULTI-seq is scalable because it uses inexpensive reagents, involves minimal sample handling, and is rapid and modular in design. MULTI-seq modularity enables any number of samples to be multiplexed with a single pair of ‘anchor’ and ‘co-anchor’ LMOs. Moreover, since LMOs are quenchable with BSA and can be incorporated during proteolytic dissociation, we anticipate that further method optimization will facilitate wash-free sample preparation workflows. When integrated with automated liquid handling, these features position MULTI-seq as a powerful technology enabling ‘screen-by-sequencing’ applications (e.g., L100048, DRUG-seq49) in multicellular systems (e.g., organoids, PBMCs, etc.).
In this study, we leveraged MULTI-seq scalability to perform a 96-plex HMEC perturbation assay, revealing noteworthy principles for future scRNA-seq sample multiplexing experiments. Specifically, we observed that responses to signaling molecules were less pronounced than responses linked to cellular composition. For instance, co-cultured MEPs and LEPs engage in TGF-β signaling that is absent in the associated monocultures. In contrast, MEPs and LEPs only exhibited pronounced transcriptional responses to the EGFR ligands AREG and EGF in these data, despite the established roles of all tested signaling molecules in mammary morphogenesis. We speculate that rich media formulations used to expand cells, such as the M87A media (-EGF) used here, likely buffer cells against microenvironmental perturbations. Thus, careful consideration of cell-type composition and media formulation will be essential to accurately interpret future scRNA-seq experiments.
Beyond its scalability, MULTI-seq improves scRNA-seq data quality in two distinct ways. First, MULTI-seq identifies doublets as cells associated with multiple sample indices. The ability to detect doublets allows for droplet-microfluidics devices to be “super-loaded”, resulting in ~5-fold improvement in cellular throughput14,21. Moreover, unlike computational doublet prediction methods32,33, MULTI-seq detects homotypic doublets and performs well on scRNA-seq data with minimal cell-type complexity. However, since computational doublet detection methods detect doublets formed from cells with shared sample barcodes, doublet detection should ideally involve a synergy of computational and molecular approaches.
Second, MULTI-seq improves scRNA-seq data quality by ‘rescuing’ cells that would otherwise be discarded by quality-control workflows utilizing RNA UMI thresholds. Such workflows are systematically biased against cell types with low RNA content39. MULTI-seq classifications provide an orthogonal metric to RNA UMIs for distinguishing low-RNA from low-quality cells. We leveraged this feature (described initially by Stoeckius et al21) to improve the quality of the PDX dataset, where MULTI-seq classifications ‘rescued’ > 90% of the sequenced neutrophils while avoiding misclassification of broken cells.
Finally, MULTI-seq is universally applicable to any sample including cells or nuclei with an accessible plasma membrane. As a result, we used the same set of MULTI-seq reagents to multiplex 15 distinct cell types or nuclei from both mice and humans. Notably, CMOs outperformed LMOs in nuclei isolation buffers containing BSA because BSA sequesters LMOs. Additionally, we anticipate that MULTI-seq is compatible with sample preservation strategies such as flash-freezing and fixation.
We leveraged all three of these features – scalability, universality, and data quality improvement – to multiplex cryopreserved primary tumors and lungs dissected from PDX mouse models at varying stages of metastatic progression. PDX sample multiplexing requires barcoding cells from (i) multiple species that may (ii) down-regulate surface epitopes commonly targeted by antibody-based multiplexing techniques (e.g., MHC-150), and (iii) have intrinsically-low viability requiring minimal sample handling. MULTI-seq successfully demultiplexed every sample, revealing novel and literature-supported immune cell responses to metastatic progression in the lung. For example, while metastasis-associated shifts in neutrophil, alveolar macrophage, and CM proportions were previously observed, we described significant shifts in interstitial macrophages, dendritic cells, and non-classical monocytes that, to our knowledge, are novel and require further experimental validation.
Moreover, we identified CM subsets that were discernible by Cd14 expression and genes with diverse effects on metastatic progression. Perplexingly, Cd14-high CMs expressing the pro-metastatic gene Thbs145 and CD14-low CMs expressing the anti-metastatic genes S100a8/9 and Wfdc2146 coexisted in metastasized lungs. Since we isolated immune cells from the whole lung in this study, we could not discern whether CD14-high and CD14-low states were spatially correlated with metastatic sites. However, MULTI-seq could be employed to spatially barcode distinct regions of a single metastatic lung, enabling direct interrogation of CM spatial heterogeneity.
In summary, MULTI-seq broadly enables users to incorporate additional layers of information into scRNA-seq experiments. In the future, we anticipate that more diverse types of information will be targeted including spatial coordinates, time-points, species-of-origin, and subcellular structures (e.g., nuclei from multinucleated cells). We also anticipate that increasing LMO membrane residency time using alternative oligonucleotide conjugate designs may enable MULTI-seq applications for non-genetic lineage tracing and/or cellular competition assays.
ONLINE METHODS (EXPERIMENTAL)
Design and synthesis of LMOs, CMOs, and sample barcode oligonucleotides:
Anchor and co-anchor LMO and CMO designs were adapted from Weber et al24. Briefly, the anchor LMO has a 5’ lignoceric acid (LA) modification with two oligonucleotide domains. The 5’ end is complimentary to the co-anchor LMO, which bears a 3’ palmitic acid (PA), and the 3’ end is complimentary to the PCR handle of the sample barcode oligonucleotide. The sample barcode was designed to have three components (as in Stoeckius et al51): (1) a 5’ PCR handle for barcode amplification and library preparation, (2) an 8 bp barcode with Hamming distance >3 relative to all other utilized barcodes, and (3) a 30bp poly-A tail necessary for hybridization to the oligo-dT region of mRNA capture bead oligonucleotides. Identically designed anchor and co-anchor CMOs are conjugated to cholesterol at the 3’ or 5’ ends via a triethylene glycol (TEG) linker and are commercially available from Integrated DNA Technologies.
| Anchor: | {LA/Chol-TEG}−5′-GTAACGATCCAGCTGTCACTTGGAATTCTCGGGTGCCAAGG-3′ |
| Co-anchor: | 5′-AGTGACAGCTGGATCGTTAC-3′-{PA/TEG-Chol} |
| Sample barcode: | 5′-CCTTGGCACCCGAGAATTCCANNNNNNNNA30-3′ |
Anchor LMO and co-anchor LMO synthesis:
Oligonucleotides were synthesized on an Applied Biosystems Expedite 8909 DNA synthesizer, as previously described (Weber et al24, Supplemental Materials).
Cell culture:
For the proof-of-concept scRNA-seq and snRNA-seq experiments, HEK293 cells, HMECs, Jurkat cells, and MEF cells were maintained at 37 °C with 5% CO2. HEK293 and MEF cells were cultured in Dulbecco’s Modified Eagle’s Medium, High Glucose (DMEM H-21) containing 4.5 g/L glucose, 0.584 g/L L-glutamine, 3.7 g/L NaHCO3, supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin (100 U/mL and 100 μg/mL, respectively). HMECs were cultured in M87A media30 with or without 24 hours of stimulation with 5 ng/mL human recombinant TGF-β (Peprotech). Jurkat cells were cultured in RPMI-1640 with 25 mM HEPES and 2.0 g/L NaHCO3 supplemented with 10% FBS and penicillin/streptomycin (100 U/mL and 100 μg/mL, respectively).
For the 96-sample HMEC experiments, fourth passage HMECs were lifted using 0.05% trypsin-EDTA for 5 minutes. The cell suspension was passed through a 45 μm cell strainer to remove any clumps. The cells were washed with M87A media once and resuspended at 107 cells/mL. The cells were incubated with 1:50 APC/Cy-7 anti-human/mouse CD49f (Biolegend, #313628) and 1:200 FITC anti-human CD326 (EpCAM) (Biolegend, #324204) antibodies for 30 minutes on ice. The cells were washed once with PBS and resuspended in PBS with 2% BSA with DAPI at 2–4 million cells/mL. Cells were sorted on BD FACSAria III. DAPI+ cells were discarded. LEPs were gated as EpCAMhi/CD49flo and MEPs were gated as EpCAMlo/CD49fhi (Fig. S7)52. Notably, this gating strategy results in trace numbers of MEPs and LEPs sorted incorrectly. HMEC sub-populations were sorted into 24-well plates such that wells contained LEPs only, MEPs only, or a 2:1 ratio of LEPs to MEPs. Sorted cell populations were cultured for 48 hours in M87A media before culturing for 72 hours in M87A media (-EGF) supplemented with different signaling molecules or signaling molecule combinations. Specifically, M87A media (-EGF) was supplemented with 100 ng/mL RANKL, 100 ng/mL WNT4, 100 ng/mL IGF-1, 113 ng/mL AREG, and/or 5 ng/mL EGF (all from Peprotech) alone or in all possible pairwise combinations. For the 96-sample HMEC technical replicate experiment, in vitro cultures were prepared as described above, except all sorted wells contained both LEPs and MEPs. Cultures were then grown in complete M87A media for 72 hours prior to isolation.
scRNA-seq sample preparation:
For the proof-of-concept experiment, cells were first treated with trypsin for 5 minutes at 37 °C in 0.05% trypsin-EDTA before quenching with appropriate cell culture media. Single-cell suspensions were then pelleted for 4 minutes at 160 rcf and washed once with PBS before suspension in 90 μL of a 200 nM solution containing equimolar amounts of anchor LMO and sample barcode oligonucleotides in PBS. Anchor LMO-barcode labeling was performed for 5 minutes on ice before 10 μL of 2 μM co-anchor LMO in PBS (for a final concentration of 200 nM) was added to each cell pool. Following gentle mixing, the labeling reaction was continued on ice for another 5 minutes before cells were washed twice with PBS, resuspended in PBS with 0.04% BSA, filtered, and pooled. The same workflow was also performed with CMOs. LMO-, CMO-, and unlabeled control cells were then loaded into three distinct 10X microfluidics lanes.
For the original 96-plex HMEC experiment, LMO labeling was performed during trypsinization in order to minimize wash steps and thereby limit cell loss and preserve cell viability. HMECs cultured in 24-well plates were labeled for 5 minutes at 37 °C and 5% CO2 in 190 μL of a 200 nM solution containing equimolar amounts of anchor LMO and sample barcode oligonucleotides in 0.05% trypsin-EDTA. 10 μL of 4 μM co-anchor LMO in 0.05% trypsin-EDTA was then added to each well (for a final concentration of 200 nM) and labeling/trypsinization was continued for another 5 minutes at 37 °C and 5% CO2 before quenching with appropriate cell culture media. A similar labeling protocol was used for the technical replicate experiment, except LMOs were incorporated once the cells were in single-cell suspension. Cells were then transferred to a 96-well plate for washing with 0.04% BSA in PBS. Finally, cells were pooled into a single aliquot, filtered through a 0.45 μm cell strainer, and counted before loading 10X microfluidics lanes.
For the PDX experiment, primary tumors and lungs were cryopreserved after dissection from triple-negative breast cancer PDX models generated in NOD-SCID gamma (NSG) mice as described previously53. The UCSF Institutional Animal Care and Use Committee (IACUC) reviewed and approved all animal experiments. On the day of the experiment, cryopreserved tissues were thawed and dissociated in digestion media containing 50 μg/mL Liberase TL (Sigma-Aldrich) and 2×104 U/mL DNase I (Sigma-Aldrich) in DMEM/F12 (Gibco) using standard GentleMacs protocols. Dissociated cells were then filtered through a 70 μm cell strainer to obtain a single-cell suspension prior to washing with PBS. Cells were then stained for 15 minutes on ice with 1:500 Zombie NIR (BioLegend, #423105) viability dye in PBS. Cells were then washed with 2% FBS in PBS prior to blocking for 5 minutes on ice with 100 μL 1:200 Fc-block (Tonbo, #70–0161-U500) in 2% FBS in PBS. After blocking, cells were stained for 45 minutes on ice with 100 μL of an antibody cocktail containing anti-mouse TER119 (FITC, ThermoFisher, #11-5921-82), anti-mouse CD31 (FITC, ThermoFisher, #11-0311-85), anti-mouse CD45 (BV450, Tonbo, #75–0451-U100), anti-mouse MHC-I (APC, eBioscience, #17-5999-82) and anti-human CD298 (PE, BioLegend, #341704). Cells were then washed with PBS prior to MULTI-seq labeling for 5 minutes on ice with 100 μL of 2.5 μM anchor LMO-barcode in PBS. 20 μL of 15 μM co-anchor LMO in PBS was added to each cell pool (for a final concentration of 2.5 μM) and labeling was continued for another 5 minutes.
Notably, we used a 10-fold greater LMO concentration for this experiment to account for increases in the total number of cells and lipophilic molecules remaining after dissociation. Following LMO labeling, cells were diluted with 100 μL of 2% FBS in PBS to ‘quench’ LMOs and washed once in 2% FBS in PBS. Finally, mCD45+ mouse immune cells and hCD298+ human metastases from dissociated primary tumors and lungs were pooled after FACS enrichment, as described previously (Lawson et al., 2015; Fig. S8, Fig. S9). Cell pools were then sequenced in a single 10X microfluidics lane.
snRNA-seq sample preparation:
For the Jurkat cell activation time-course, 2×105 Jurkat cells were added to 8 wells of a 12-well plate and treated with 10 ng/μL phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich #P8139) and 1.3 μM ionomycin (Sigma-Aldrich #I0634) at 15 min, 30 min, 1 hr, 2 hr, 4 hr, 6 hr, or 24 hr prior to barcoding with LMOs. A single well of Jurkat cells were left untreated. HEK293 and MEF cells were cultured as described above. Nuclei were isolated from cells using a protocol adapted from 10X Genomics. Briefly, suspensions of HEK293, MEF, or treated Jurkat cells were washed once with PBS, pelleted at 160 rcf (HEK293, MEFS) or 300 rcf (Jurkat) for 4 min at 4 °C and suspended in chilled lysis buffer (0.5% Nonidet P40 Substitute, 10 mM Tris-HCl, 10 mM NaCl, and 3 mM MgCl2 in milliQ water) to a density of 2.5 × 106 cells/mL. Lysis proceeded for 5 minutes on ice, after which the lysate was pelleted (500 rcf, 4 °C, 4 minutes) and washed three times in chilled resuspension buffer (2% BSA in PBS). Nuclei were then diluted to a concentration of ~106 nuclei/mL prior to LMO or CMO labeling. HEK293 and MEF cells were each divided into two samples and labeled with LMOs or CMOs (500 nM in resuspension buffer) using the same procedure as described for live cells (presence of BSA during labeling is the lone alteration as it is required to prevent nuclei clumping). Each Jurkat sample was labeled with LMOs, alone. Each sample was washed 3X in 1mL resuspension buffer (500 rcf, 4 °C, 4 min). The four LMO- and CMO-labeled HEK293 and MEF samples were pooled in equal portions and, separately, Jurkat samples were pooled in equal proportions. These final two samples were combined in a 1:1 ratio and sequenced on a single 10X microfluidics lane.
scRNA-seq and snRNA-seq library preparation:
Sequencing libraries were prepared using a custom protocol based on the 10X Genomics Single Cell V2 and CITE-seq51 workflows. Briefly, the 10X workflow was followed up until cDNA amplification, where 1 μL of 2.5 μM MULTI-Seq additive primer was added to the cDNA amplification master mix:
| MULTI-seq additive primer: | 5′-CCTTGGCACCCGAGAATTCC-3′ |
This primer increases barcode sequencing yield by enabling the amplification of barcodes that successfully primed reverse transcription on mRNA capture beads but were not extended via template switching (Fig. S10C). Notably, the MULTI-seq additive primer was erroneously excluded during the proof-of-concept snRNA-seq library preparation, and nuclei were still able to be robustly classified. Following amplification, barcode and endogenous cDNA fractions were separated using a 0.6X SPRI size selection. The endogenous cDNA fraction was then processed according to the 10X workflow until next-generation sequencing (NGS) with the following formats:
| Dataset | NGS Format |
|---|---|
| Proof-of-concept (scRNA-seq) | 2x HiSeq 4000 |
| Proof-of-concept (snRNA-seq) | NovaSeq (20%) |
| HMEC | NovaSeq (100%) |
| HMEC (technical replicate) | NovaSeq (5%) |
| PDX | NovaSeq (70%) |
To prepare the barcode fraction for NGS, contaminating oligonucleotides remaining from cDNA amplification were first removed using an established small RNA enrichment protocol (Beckman Coulter). Specifically, we increased the final SPRI ratio in the barcode fraction to 3.2X reaction volumes and added 1.8X reaction volumes of 100% isopropanol (Sigma-Aldrich). Beads were then washed twice with 400 μL of 80% ethanol and allowed to air dry for 2–3 minutes before elution with 50 μL of Buffer EB (Qiagen, USA). Eluted barcode cDNA was then quantified using QuBit before library preparation PCR (95 °C, 5’; 98 °C, 15”; 60 °C, 30”; 72 °C, 30”; 8 cycles; 72 °C, 1’; 4 °C hold). Each reaction volume was a total of 50 μL containing 26.25 μL 2X KAPA HiFi HotStart master mix (Roche), 2.5 μL of 10 μM TruSeq RPIX primer (Illumina), 2.5 μL of 10 μM TruSeq Universal Adaptor primer (Illumina), 3.5 ng barcode cDNA, and nuclease-free water.
TruSeq RPIX:
5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′
TruSeq P5 Adaptor:
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′
Following library preparation PCR, remaining sequencing primers and contaminating oligonucleotides were removed via a 1.6X SPRI clean-up. Representative Bioanalyzer traces at different stages of the MULTI-seq library preparation workflow are documented in Fig. S10. Barcode libraries were sequenced using the NGS formats documented in Table S2. Notably, sequencing reads predominantly aligned to the barcode reference sequences, and resulted in high SNRs with low rates of duplicated UMIs, suggesting that barcode libraries were not sequenced to saturation for any of the presented experiments.
ONLINE METHODS (COMPUTATIONAL)
Expression library pre-processing:
Expression library FASTQs were pre-processed using CellRanger (10X Genomics) and aligned to the hg19 (proof-of-concept scRNA-seq, HMEC), concatenated mm10-hg19 (PDX), or concatenated mm10-hg19 pre-mRNA (proof-of-concept snRNA-seq) reference transcriptomes. When multiple 10X lanes were sequenced in an experiment, CellRanger aggregate was used to perform read-depth normalization.
Cell/Nuclei calling:
For the proof-of-concept scRNA-seq, snRNA-seq and HMEC technical replicate experiments, cell-associated barcodes were defined using CellRanger. For the original 96-plex HMEC experiment, cells were defined as cell barcodes (1) associated with >= 600 total RNA UMIs that (2) were successfully classified during MULTI-seq sample classification workflow. We manually selected 600 RNA UMIs as a threshold in order to exclude low-quality cell barcodes. For the PDX experiment, we defined cells as barcodes (1) associated with >= 100 total RNA UMIs that (2) were successfully classified during the MULTI-seq sample classification workflow (Supplemental Materials).
Expression library analysis:
Following pre-processing and cell/nuclei calling, RNA UMI count matrices were prepared for analysis using the ‘Seurat’ R package, as described previously54,55. Briefly, genes expressed in fewer than 3 cells were discarded before the percentage of reads mapping to mitochondrial genes (%Mito) was computed for each cell. Outlier cells with elevated %Mito were visually defined and discarded. Data was then log2-transformed, centered, and scaled before variance due to %Mito and the total number of RNA UMIs were regressed out. Highly variable genes were then defined for each dataset by selecting mean expression and dispersion thresholds resulting in ~2000 total genes. These variable genes were then used during PCA, and statistically-significant PCs were defined by PC elbow plot inflection point estimation. Significant PCs were then utilized for unsupervised Louvian clustering and dimensionality reduction with t-SNE56.
Following pre-processing, differential gene expression analysis was performed using the ‘FindMarkers’ command in ‘Seurat’, with ‘test.use’ set to ‘bimod’57 and log fold-change thresholds set in a context-dependent fashion (Supplemental Materials). Other dataset-specific analyses are discussed in the Supplemental Materials. Dataset-specific ‘Seurat’ pre-processing parameters:
| Dataset | PCs | Variable gene dispersion threshold | Variable gene expression threshold |
|---|---|---|---|
| Proof-of-concept, live cells | 8 | 0.5 | 0.025 |
| Proof-of-concept, human nuclei | 8 | 0.5 | 0.0125 |
| Proof-of-concept, mouse nuclei | 6 | 0.5 | 0.0125 |
| Proof-of-concept, Jurkat nuclei | 10 | 0.5 | 0.0125 |
| 96-Plex HMEC, all cells | 8 | 0.65 | 0.05 |
| 96-Plex HMEC, all LEPs | 9 | 0.4 | 0.05 |
| 96-Plex HMEC, all MEPs | 10 | 0.75 | 0.05 |
| 96-Plex HMEC, resting LEPs | 8 | 0.4 | 0.05 |
| 96-Plex HMEC, resting MEPs | 6 | 1.0 | 0.05 |
| PDX, mouse immune cells | 15 | 0.75 | 0.05 |
| PDX, mouse lung immune cells | 14 | 0.7 | 0.05 |
| PDX, classical monocytes | 10 | 0.53 | 0.05 |
| PDX, late-stage classical monocytes | 6 | 0.65 | 0.05 |
Barcode library pre-processing:
Raw barcode library FASTQs were converted to barcode UMI count matrices using custom scripts leveraging the ‘ShortRead’58 and ‘stringdist’59 R packages (Fig. S3). Briefly, raw FASTQs were first parsed to discard reads where the first 16 bases of R1 did not perfectly match any of the cell barcodes associated a pre-defined list of cell barcodes. Second, reads where the first 8 bases of R2 did not align with < 1 mismatch to any reference barcode were discarded. Third, reads were binned by cell barcodes and duplicated UMIs were identified as reads where bases 17–26 of R2 exactly matched. Finally, reference barcode alignment results were then parsed to remove duplicated UMIs before being converted into a final barcode UMI count matrix.
Barcode library sequencing statistics:
MULTI-seq barcode library sequencing statistics were computed for classified singlets in all datasets presented in this study. SNR was computed for every cell by finding the quotient of the top two most abundant barcodes. Mean SNRs among all singlets for each dataset presented in this study are documented in Table S2. The alignment rate was defined as the proportion of singlet-associated sequencing reads where the first 8 bases of R2 aligned with < 1 mismatch to any reference barcode.
MULTI-seq sample classification:
MULTI-seq barcode UMI count matrices were used to classify cells into sample groups via a workflow inspired by previous scRNA-seq multiplexing approaches15,16,21 (Fig. S3). First, raw barcode reads were log2-transformed and mean-centered. The presence of each barcode was then visually inspected by performing t-SNE on the normalized barcode count matrix, as implemented in the ‘Rtsne’ R package with ‘initial_dims’ set to the total number of barcodes56. Missing barcodes (observed only for the 96-plex HMEC experiment) were discerned as those lacking any enrichment in barcode space and were removed.
Next, the top and bottom 0.1% of values for each barcode were excluded and the probability density function (PDF) for each barcode was defined by applying the ‘approxfun’ R function to Gaussian kernel density estimations produced using the ‘bkde’ function from the ‘KernSmooth’ R package60. We then sought to classify cells according to the assumption that groups of cells that are positive and negative for each barcode should manifest as local PDF maxima15,16. To this end, we computed all local maxima for each PDF and defined negative and positive maxima as the most frequent and highest local maxima, respectively. Notably, this strategy assumes that truly-barcoded cells will have the highest abundance for any given barcode, and that no individual sample group will have more members than the sum of all other groups.
With these positive and negative approximations in hand, we next sought to define barcode-specific UMI thresholds. To find the best inter-maxima quantile for threshold definition (e.g., an inter-maxima quantile of 0.5 corresponds to the mid-point), we iterated across 0.02-quantile increments and chose the value that maximized the number of singlet classifications. Sample classifications were then made using these barcode-specific UMI thresholds by discerning which thresholds each cell surpasses, with doublets being defined as cells surpassing > 1 threshold21. Negative cells (i.e., cells surpassing 0 thresholds) were then removed, and this procedure was repeated until all cells were classified as singlets or doublets. Subsets of negative cells could then be reclassified using semi-supervised learning21, where singlets defined during the initial workflow are used to initialize cluster centers during k-means clustering of negative cells (Supplemental Materials).
Statistical Tests:
Statistically-significant TGFBI expression enrichment amongst TGF-β-stimulated and unstimulated HMECs in the proof-of-concept scRNA-seq experiment was assessed using the Wilcoxon rank-sum test (two-sided, n = 1,950 cells). Statistically-significant TGFBI expression enrichment amongst LEPs and MEPs grouped according to signaling molecule exposure was assessed using the Wilcoxon rank-sum test (two-sided, n = 32 signaling molecule condition groups). Differentially expressed genes between clusters in all datasets were defined using the likelihood-ratio test for single cell gene expression57 with Bonferroni multiple comparisons adjustment. Statistically-significant changes in lung immune cell type proportions during metastatic progression were assessed using the two-proportion z-test with Bonferroni multiple comparisons adjustment (n = 44 tumor-stage/cell type groups).
DATA AVAILABILITY
Raw gene expression and barcode count matrices were uploaded to the Gene Expression Omnibus (GSE…) along with pertinent metadata.
CODE AVAILABILITY
R implementations of the MULTI-seq sample classification and barcode pre-processing pipelines are available in the ‘deMULTIplex’ R package, and can be downloaded at https://github.com/chris-mcginnis-ucsf/MULTI-seq.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by grants from the Department of Defense Breast Cancer Research Program (W81XWH-10-1-1023 and W81XWH-13-1-0221), NIH (U01CA199315 and DP2 HD080351-01), the NSF (MCB-1330864), and the UCSF Center for Cellular Construction (DBI-1548297), an NSF Science and Technology Center. Z.J.G is a Chan-Zuckerberg BioHub Investigator. D.M.P. is supported by the NIGMS of the National Institutes of Health (F32GM128366). L.M.M is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2239-15). J.W. and M.Y.H. are supported by EMBO long-term postdoctoral fellowships (ALTF-159-2017 and ALTF-1193-2015, respectively). J.L.H. is supported by an NSF GRFP award. We thank M. Thomson (Pasadena, CA) for insightful discussions. We thank the UCSF Flow Core (NIHS10 1S10OD021822-01) and M. Owyong, S. Liu and C. Diadhiou (San Francisco, CA) for their technical support.
Footnotes
DECLARATION OF INTERESTS
Z.J.G., E.D.C., D.M.P., and C.S.M. have filed patent applications related to the MULTI-seq barcoding method. The contents of this manuscript are solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Ramsköld D, Luo S, Wang Y, et al. Full-Length mRNA-Seq from single cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012; 30(8): 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012; 2(3):666–73. [DOI] [PubMed] [Google Scholar]
- 3.Gierahn TM, Wadsworth MH 2nd, Hughes TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017; 14(4):395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao J, Packer JS, Ramani V, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017; 357(6352):661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg AB, Roco CM, Muscat RA, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science. 2018; 360(6385):176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macosko EZ, Basu A, Satija R, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. 2015. Cell; 161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. 2015. Cell; 161(5):1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017; 14(10):955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabula Muris Consortium. Single-cell transcriptomic characterization of 20 organs and tissues from individual mice creates a Tabula Muris. Nature. 2018; 562(7727):367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regev A, Teichmann SA, Lander ES, et al. The Human Cell Atlas. Elife. 2017; 6 pii: e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018; 360(6392):981–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordovas-Montanes J, Dwyer DF, Nyquist SK, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. 2018; 560(7720):649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HM, Subramaniam M, Targ S, et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol. 2018; 36(1):89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dixit A, Parnas O, Li B, et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. 2016; 167(7):1853–66.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson B, Norman TM, Jost M, et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016; 167(7):1867–82.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaitin DA, Weiner A, Yofe I, et al. Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell. 2016; 167(7):1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- 18.Aarts M, Georgilis A, Beniazza M, et al. Coupling shRNA screens with single-cell RNA-seq identifies a dual role for mTOR in reprogramming-induced senescence. Genes Dev. 2017; 31(20):2085–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin D, Lee W, Lee JH, Bang D. Multiplexed single-cell RNA-seq via transient barcoding for drug screening. 2018. Preprint: bioRxiv doi: 10.1101/359851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo C, Biddy BA, Kamimoto K, Kong W, Morris SA. CellTag Indexing: a genetic barcode-based multiplexing tool for single-cell technologies. 2019. Preprint: bioRxiv doi: 10.1101/335547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoeckius M, Zheng S, Houck-Loomis B, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018; 19:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gehring J, Park JH, Chen S, Thomson M, Pachter L. Highly Multiplexed Single-Cell RNA-seq for Defining Cell Population and Transcriptional Spaces. 2018. Preprint bioRxiv doi: 10.1101/315333. [DOI] [Google Scholar]
- 23.Gaublomme JT, Li B, McCabe C, et al. Nuclei multiplexing with barcoded antibodies for single-nucleus genomics. 2018. Preprint bioRxiv doi: 10.1101/476036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber RJ, Liang SI, Selden NS, Desai TA, Gartner ZJ. Efficient targeting of fatty-acid modified oligonucleotides to live cell membranes through stepwise assembly. Biomacromolecules. 2014; 15(12):4621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol. 2019; 30(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coutelier JP, Kehrl JH, Bellur SS, et al. Binding and functional effects of thyroid stimulating hormone on human immune cells. J Clin Immunol. 1990;10(4):204–10. [DOI] [PubMed] [Google Scholar]
- 27.Jeffrey KL, Brummer T, Rolph MS, et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat Immunol. 2006; 7(3):274–83. [DOI] [PubMed] [Google Scholar]
- 28.Ziegler SF, Ramsdell F, Alderson MR. The activation antigen CD69. Stem Cells. 1994; 12(5):456–65. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman J, Fan Z. Nuclear war: the granzyme A-bomb. Curr Opin Immunol. 2003; 15(5):553–9. [DOI] [PubMed] [Google Scholar]
- 30.Garbe JC, Bhattacharya S, Merchant B, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009; 69(19):7557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brisken C. Progesterone signalling in breast cancer: a neglected hormone coming into the limelight. Nat Rev Cancer. 2013; 13(6):385–96. [DOI] [PubMed] [Google Scholar]
- 32.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. 2019. Cell Systems (in press). doi: 10.1101/352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolock SL, Lopez R, Klein AM. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. 2019. Cell Sysmtes (in press). doi: 10.1101/357368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chitale D, Gong Y, Taylor BS, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009; 28(31):2773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fearon AE, Carter EP, Clayton NS, et al. PHLDA1 Mediates Drug Resistance in Receptor Tyrosine Kinase-Driven Cancer. Cell Rep. 2018; 22(9):2469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage P, Blanchet-Cohen A, Revil T, et al. A Targetable EGFR-Dependent Tumor-Initiating Program in Breast Cancer. Cell Rep. 2017; 21(5):1140–1149. [DOI] [PubMed] [Google Scholar]
- 37.DeRose YS, Wang G, Lin YC, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011; 17(11):1514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang K, Sun X, Chen Y, Shen Y, Jarvis JN. RNA sequencing from human neutrophils reveals distinct transcriptional differences associated with chronic inflammatory states. BMC Med Genomics. 2015; 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lun ATL, Riesenfeld S, Andrews T, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 2019; 20:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reyfman PA, Walter JM, Joshi N, et al. Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med. 2018. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jablonska J, Lang S, Sionov RV, Granot Z. The regulation of pre-metastatic niche formation by neutrophils. Oncotarget. 2017; 8(67):112132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma SK, Chintala NK, Vadrevu SK, Patel J, Karbowniczek M, Markiewski MM. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J Immunol. 2015; 194:5529–38. [DOI] [PubMed] [Google Scholar]
- 43.Condamine T, Ramachandran I, Youn J, Gabrilovich DI. Regulation of Tumor Metastasis by Myeloid-derived Suppressor Cells. Annu Rev Med. 2015; 66:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamura T, Doughty-Shenton D, Cassetta L, et al. Monocytes Differentiate to Immune Suppressive Precursors of Metastasis-Associated Macrophages in Mouse Models of Metastatic Breast Cancer. Front Immunol. 2018; 8:2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Catena R, Bhattacharya N, El Rayes T, et al. Bone marrow-derived Gr1+ cells can generate a metastasis-resistant microenvironment via induced secretion of thrombospondin-1. Cancer Discov. 2013; 3:578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouzounova M, Lee E, Piranlioglu R, et al. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun. 2017; 8:14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabavi S, SChmolze D, Maitituoheti M, Malladi S, Beck AH. EMDomics: a robust and powerful method for the identification of genes differentially expressed between heterogeneous classes. Bioinformatics. 2016; 32(4):533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell. 2017; 171(6):1437–52.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye C, Ho DJ, Neri M, Yang C, Kulkarni T, Randhawa R, et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat Commun. 2018; 9(1):4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero JM, Jiménez P, Cabrera T, et al. Coordinated downregulation of the antigen presentation machinery and HLA class I/beta2-microglobulin complex is responsible for HLA-ABC loss in bladder cancer. Int J Cancer. 2005; 113(4):605–10. [DOI] [PubMed] [Google Scholar]
- 51.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017; 14(9):865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009; 15(8):907–13. [DOI] [PubMed] [Google Scholar]
- 53.Lawson DA, Bhakta NR, Kessenbrock K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015; 526(7571):131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Satija R, Ferrel JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015; 33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018; doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Maaten LJP. Accelerating t-SNE using Tree-Based Algorithms. JMLR. 2014; 15:3221–45. [Google Scholar]
- 57.McDavid A, Finak G, Chattopadyay PK, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics. 2013; 29(4):461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. ShortRead: a Bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009; 25:2607–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Loo M. The stringdist package for approximate string matching. The R Journal. 2014; 6:111–22. [Google Scholar]
- 60.Wand MP & Jones MC. Kernel Smoothing Monographs on Statistics and Applied Probability. Chapman & Hall, 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw gene expression and barcode count matrices were uploaded to the Gene Expression Omnibus (GSE…) along with pertinent metadata.