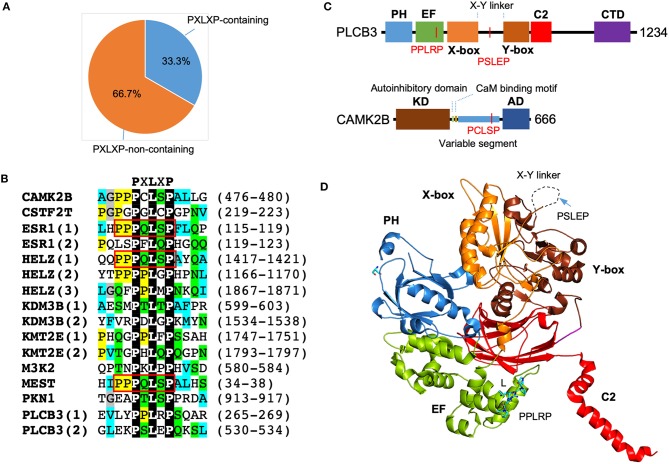
Figure 4.
Enrichment of PXLXP motif in SMYD3 interacting proteins. (A) The percentage of PXLXP containing proteins in SMYD3 interactors. (B) Sequence alignment of SMYD3 interacting proteins at PXLXP motif. Conserved motif-residues are shown as white on black. Other residues are colored according to ClustalX grouping scheme (Larkin et al., 2007): proline (yellow), glycine (gray), small or hydrophobic (C, A, V, L, I, M, F, W) (cyan), hydroxyl or amine (S, T, N, Q) (green), charged (D, E, R, K), and histidine or tyrosine (H, Y). Residues are colored only if the percentage of residues from a group is larger than 25%. Numbering at the right end of the sequences indicates the start and end of PXLXP motif. Red boxes indicate identical PQLSP motif. (C) Domain structure of PLCB3 and CAMK2B. In PLCB3, PH, pleckstrin homology domain; EF, EF-hands domain; X-box, catalytic X domain; Y-box, catalytic Y domain; C2, C2 domain; CTD, C-terminal domain. In CAMK2B, KD, kinase domain; AD, self-association domain; CaM, calmodulin. Red lines indicate the positions of PXLXP motifs. (D) Ribbon diagram of PLCB3 structure (PDB code: 3OHM). PLCB3 domains are colored according to the scheme in (C). Dash line indicates disordered X-Y linker. PPLRP motif is depicted by sticks. PSLEP motif is indicated by an arrow.