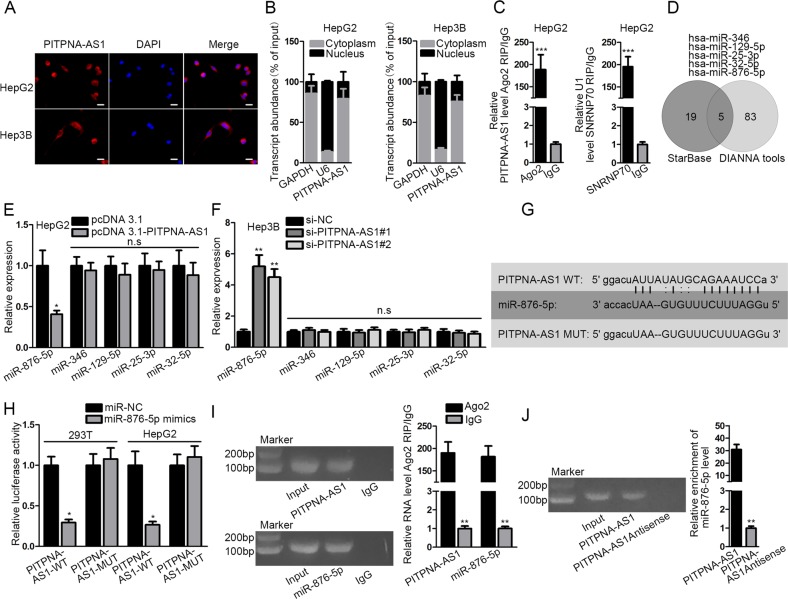
Fig. 4. MiR-876-5p was significantly targeted by PITPNA-AS1.
a, b The location of PITPNA-AS1 in HCC cells was tested by FISH and subcellular fractionation assays. c The involvement of PITPNA-AS1 in RISC was assessed by Ago2-RIP assay. d Five miRNAs that could bind with PITPNA-AS1 were predicted using starBase and DIANA tools. e, f The levels of five miRNAs were analyzed with qRT-PCR in two HCC cells after PITPNA-AS1 overexpression or silencing. g The binding sites between PITPNA-AS1 (wild-type or mutant) and miR-876-6p were exhibited. h Luciferase activity of PITPNA-AS1-WT or PITPNA-AS1-MUT was assessed in 293T or HepG2 cells transfected with miR-876-5p mimics or miR-NC. i The enrichment of PITPNA-AS1 and miR-876-5p in RISC was identified with Ago2-RIP assay. j The interaction between PITPNA-AS1 and miR-876-5p was determined by RNA pull-down assays. *P < 0.05, **P < 0.01, ***P < 0.001 indicated statistically significant differences. n.s no significance, PITPNA-AS1 phosphatidylinositol transfer protein alpha antisense RNA 1, HCC hepatocellular carcinoma, RISC RNA-induced silencing complex, qRT-PCR quantitative real time polymerase chain reaction