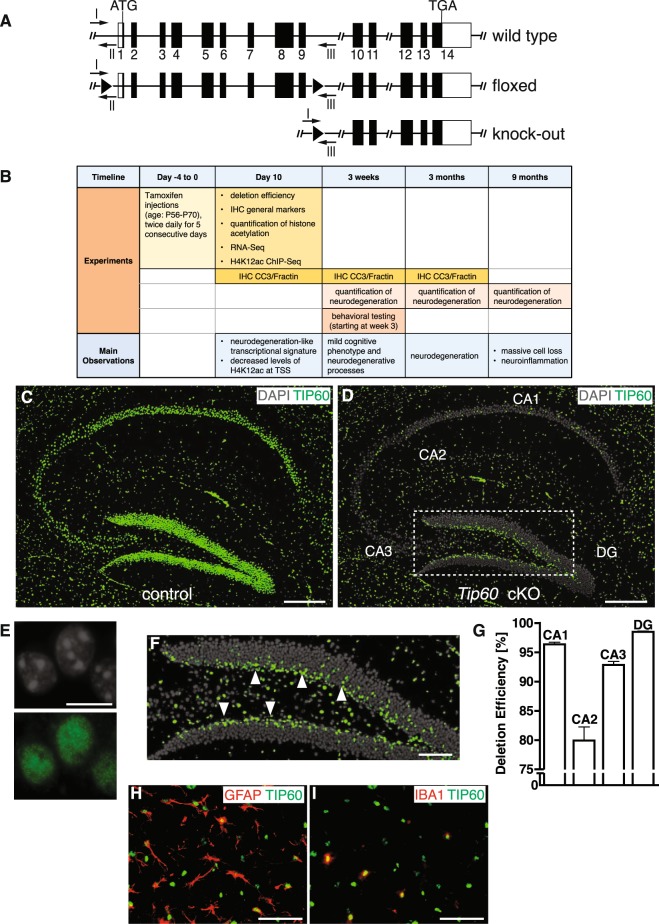
Figure 1.
Deletion of Tip60 in excitatory forebrain neurons of adult mice. (A) Tip60 alleles for wild type, floxed, and knock-out. Protein coding exons are shown in black. LoxP sites are indicated with black triangles. Genotyping primers I, II and III are indicated with arrows. (B) Time points when experiments were conducted and main observations. The last day of tamoxifen injections is defined as day 0. (C) Ubiquitous nuclear TIP60 signal in the hippocampus of control mice at day 10 after the last tamoxifen injection. (D) In Tip60 cKO most of the TIP60 signal in the principal cell layers is absent at day 10 after the last tamoxifen injection. (E) TIP60 signal is nuclear. TIP60 and DAPI staining in single hippocampal nuclei are shown. (F) Marked area from (D) showing TIP60-positive nuclei in the subgranular zone (arrowheads), a region lacking CRE activity. (G) Quantification of deletion efficiencies in CA1, CA2, CA3 and DG in Tip60 cKO mice at day 10 after tamoxifen injections, normalized to the total number of DAPI positive nuclei (n = 4, 4 sections per animal). Error bars represent SEM. (H,I) Pictures of triple labeling of TIP60, GFAP and IBA1 in the stratum lacunosum moleculare of the CA1 region in a Tip60 cKO mouse. (H) Shows TIP60 and GFAP, (I) TIP60 and IBA1. Scale bars: 250 µm (C,D), 10 µm (E), 100 µm (F), 50 µm (H,I). Abbreviations: CA1, hippocampal subfield CA1; CA2, hippocampal subfield CA2; CA3, hippocampal subfield CA3; DG, dentate gyrus; IHC, immunohistochemistry; CC3, cleaved Caspase 3.