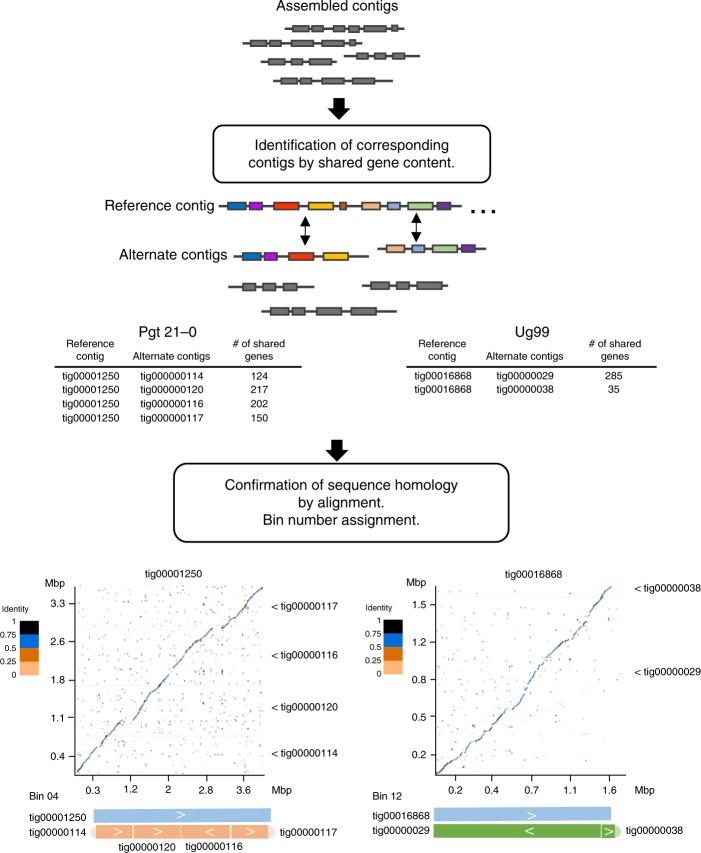
Fig. 1.
Strategy to identify homologous contigs in genome assemblies by gene synteny. To detect shared content, Pgt gene models21 (grey and coloured boxes) were aligned to the genome assemblies and the contig positions of the top two hits of each gene were recorded. Contigs containing at least five shared genes were considered as potential haplotype pairs. Sequence collinearity between contigs was assessed by alignment, and homologous matching contigs were assigned to bins. Examples shown are for Bin04 and Bin12 from Pgt21-0 and Ug99, respectively