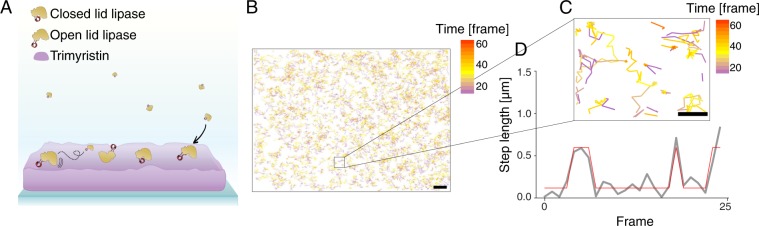
Figure 1.
Experimental setup to track individual lipase enzymes on triglyceride substrate layers using Total Internal Reflection microscopy. (A) Representation (not to scale) of triglyceride layer labeled with DOPE-ATTO-655 and Alexa Fluor 488 labeled TLL lipases displaying diffusion, multiple potential binding or interaction modes and initial lipase to substrate binding. (B) Overlay of typical temporal trajectories of individual lipases displaying lateral diffusion on trimyristin surfaces. Enzyme tracks are color-coded according to observation time. Briefly, the color code display time for a given trajectory in frames observed, purple is after enzyme binding, yellow at intermediate and red after longer observation times. Data from 100 frames are displayed for clarity, Scale bar 5 µm. (C) Closeup of traces reveals heterogeneities within diffusional behavior such as total immobilization, periods of slow diffusion or fast diffusion. Color-code as for B. Scale bar 2 µm. (D) Typical step length trace of an enzyme displaying reversible transition from initial high mobility to a low mobility state and the corresponding idealized traces found by HMM analysis.