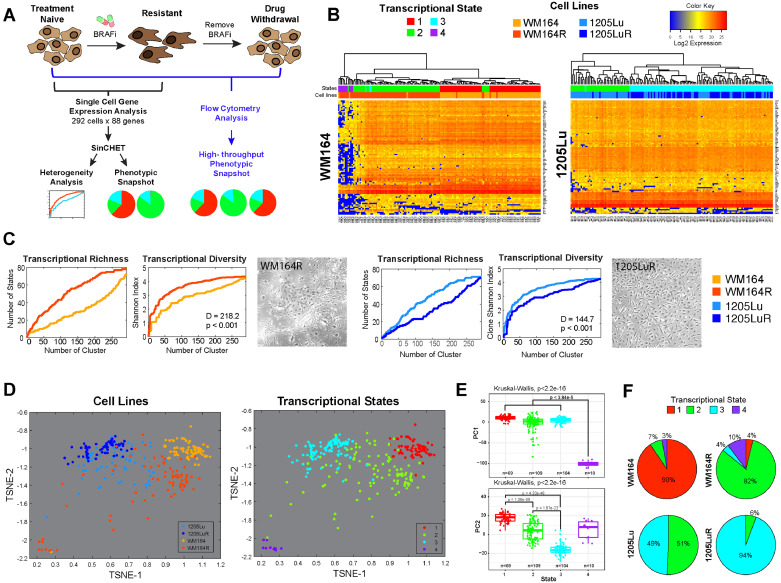
Fig. 1.
Defining the transcriptional diversity of melanoma. A. Overview of the experimental workflow. B. Single cell mRNA analysis of BRAF inhibitor naïve and treated WM164 and 1205Lu melanoma cells identifies 4 different transcriptional subpopulations. Heatmaps of gene expression data of 88 genes (y-axis) for individual cells (x-axis) of treatment-naive WM164 and 1205Lu and their drug-resistant (R) counterparts. Colour-coding under the dendogram depicts the transcriptional state and the cell line each cell belongs to. C. BRAF inhibition leads to either an increase or a decrease in transcriptional diversity. Shannon heterogeneity analyses of the single cell data from Fig. 1B reveals WM164 to become more diverse on therapy and the 1205Lu cells to become less diverse. Transcriptional richness is a measure of distinct transcriptional states present. Graphs show the number of distinct transcriptional states, as well as the Shannon Diversity Index, at different heights along the gene expression dendogram from Fig. 1B. D. t-SNE analysis demonstrates the relationship between drug-naïve and treated melanoma cell lines and the 4 transcriptional states. Data points show the individual cells and their orientation in transcriptional space. Note that drug-resistant (WM164R) cells are the most transcriptionally diverse. States #1, #3 and #4 are quite distinct with #2 showing overlap with #1 and #3. E. Four transcriptional states are statistically distinct. Principal component analysis and Kruskal-Wallis test performed comparing all cells in each transcriptional state highlights strong differences in gene expression among the four states. F. WM164 and 1205Lu cells show different transcriptional landscapes, which change under chronic BRAF inhibitor treatment. Pie charts are derived from data shown in Fig. 1B using SinCHet software analysis and show the percentage of cells in each transcriptional state.