Fig. 2.
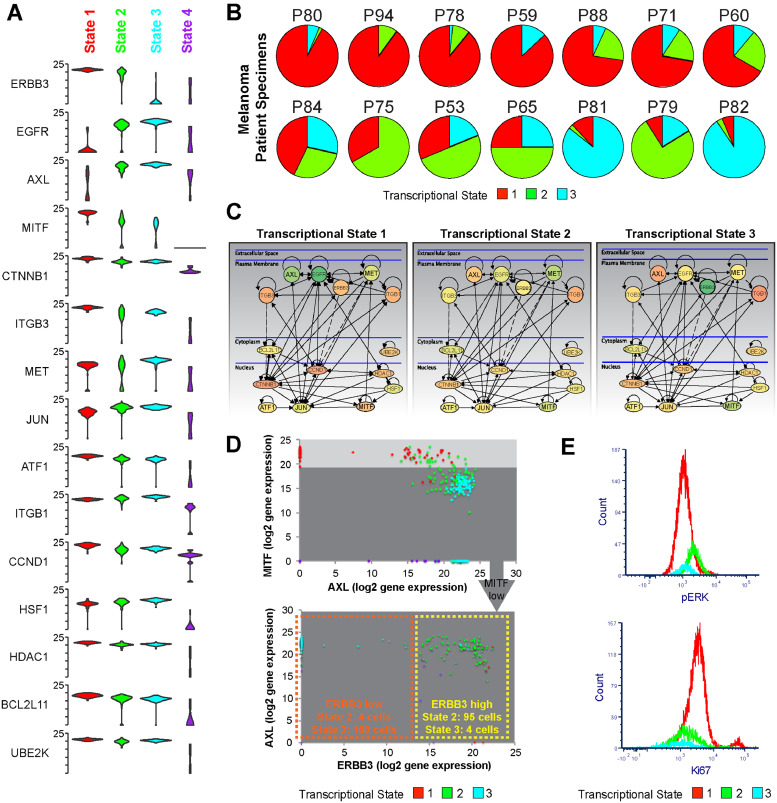
Gene level analysis identifies unique transcriptional profiles for each transcriptional state. A. Violin plots showing expression of each of the top 15 differentially expressed mRNAs clustered by transcriptional state. Note higher Axl and c-JUN in States #2 and #3, Cyclin D1 (CCND1), MITF in State #1, ERBB3 in States #1 and #2 B. Melanoma patient samples exhibit a diverse array of transcriptional state compositions. Data shows heterogeneity analysis of single cell RNA-Seq data from ref. (22) of melanoma patient specimens, based upon the 88 genes from our panel. C. major signalling differences among the three main transcriptional states. Metacore interactome analysis of the top 15 differentially expressed genes among the three transcriptional states illustrates rewiring of major signalling among the subpopulations. Node colour denotes mean level of mRNA expression detected (green = lower expression, red = higher expression). D. MITF-High/Axl-low and MITF-low/Axl-high expression signatures are not binary states. Dot plots showing the relative expression of MITF, AXL and ERBB3 in all cells derived from data in Fig. 1B show the varied distribution of MITF/AXL expression and the need to examine additional markers (such as ERBB3) for a more complete analysis of the transcriptionally heterogeneous subpopulations. E. Transcriptional State #2 has the highest pERK levels and Transcriptional State #1 shows the highest proliferative capacity. Comparison of pERK and Ki67 staining by flow cytometry between transcriptional States #1, 2 and 3. Data shows expression of phospho-ERK in each transcriptional state resolved by the gating shown in Supplemental Fig. 4 in WM164 and WM164R cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)