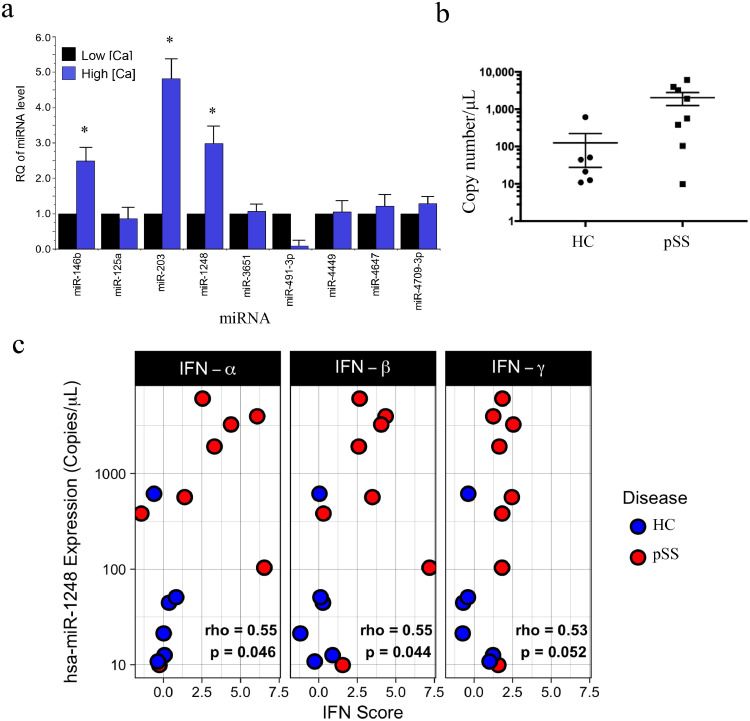
Fig. 1.
Validation of differentially expressed miRNAs in phSG cells and in SS patients. (a) Total RNAs were isolated from phSG cells maintained either in KGM-L or KGM-H medium and used for RT-qPCR with indicated TaqMan probes. Data are average of three separate experiments with mean ± S.E. and presented as a fold change over each respective low and high calcium sample (* p < .05, Student's t-test). (b) Expression level of miR-1248 in MSG of Healthy Controls (n = 6, circle) and SS subjects (n = 8, square) measured by digital PCR as copies/μl. (c) Total RNA-seq and digital PCR assays were conducted with total RNAs from healthy control (n = 6, blue) and SS patients (n = 8, red) biopsies for whole transcriptome analysis and miR-1248 copy number determination, respectively. Data are presented as the copy number of miR-1248 vs. interferon score of type I and II IFN. The interferon score is the average number of standard deviations from the mean expression level of IFN stimulated genes in healthy controls (IFN-α: IFIT1, IFI44, EIF2AK2; IFN-β: ISG15, OAS1, IFIH1, RSAD2, IFI6; IFN-γ: IRF1, GBP1, SERPING1). Each panel is labeled with the Spearman's rho value for the correlation between copy number and IFN score of all samples in the panel and the p-value from a Spearman rank test. HC: healthy control; pSS: Sjögren's syndrome patient. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)