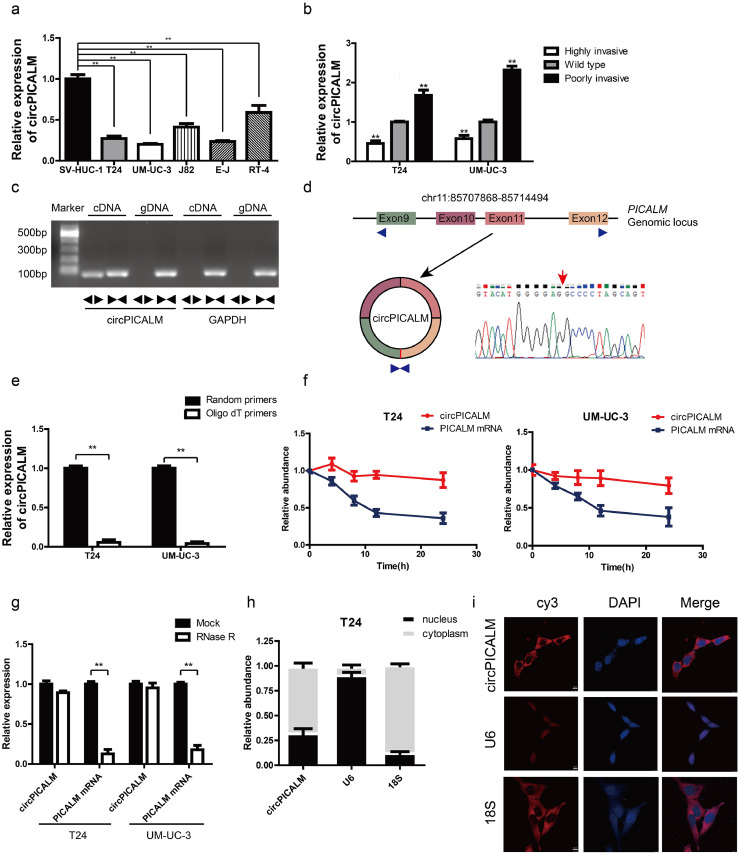
Fig. 1.
Characteristic of circPICALM.
a. Relative expression of circPICALM in SV-HUC-1, T24, UM-UC-3, J82, E-J, and RT-4 cells measured by RT-PCR. b. Relative abundance of circPICALM in our previously established invasion cell models. c. PCR products of divergent or convergent primers and T24 cDNA or gDNA were validated by gel electrophoresis. d. Schematic illustration of formation of circPICALM. The junction site was proved by Sanger sequence and indicated by red arrow. e. RT-PCR analysis of circPICALM expression in random primers or oligo dT primers reverse transcription systems. f. Relative abundance of circPICALM and linear PICALM in T24 and UM-UC-3 cells treated with 5μg/ml actinomycin D. RNAs were measured via RT-PCR at the indicated time. g. RT-PCR results of circPICALM and linear PICALM in BC cells treated with or without RNase R. h. Subcellular distribution of circPICALM was detected by nuclear mass separation assay in T24 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. i. Subcellular location of circPICALM was illustrated by FISH in UM-UC-3 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. Scale bar, 10 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).