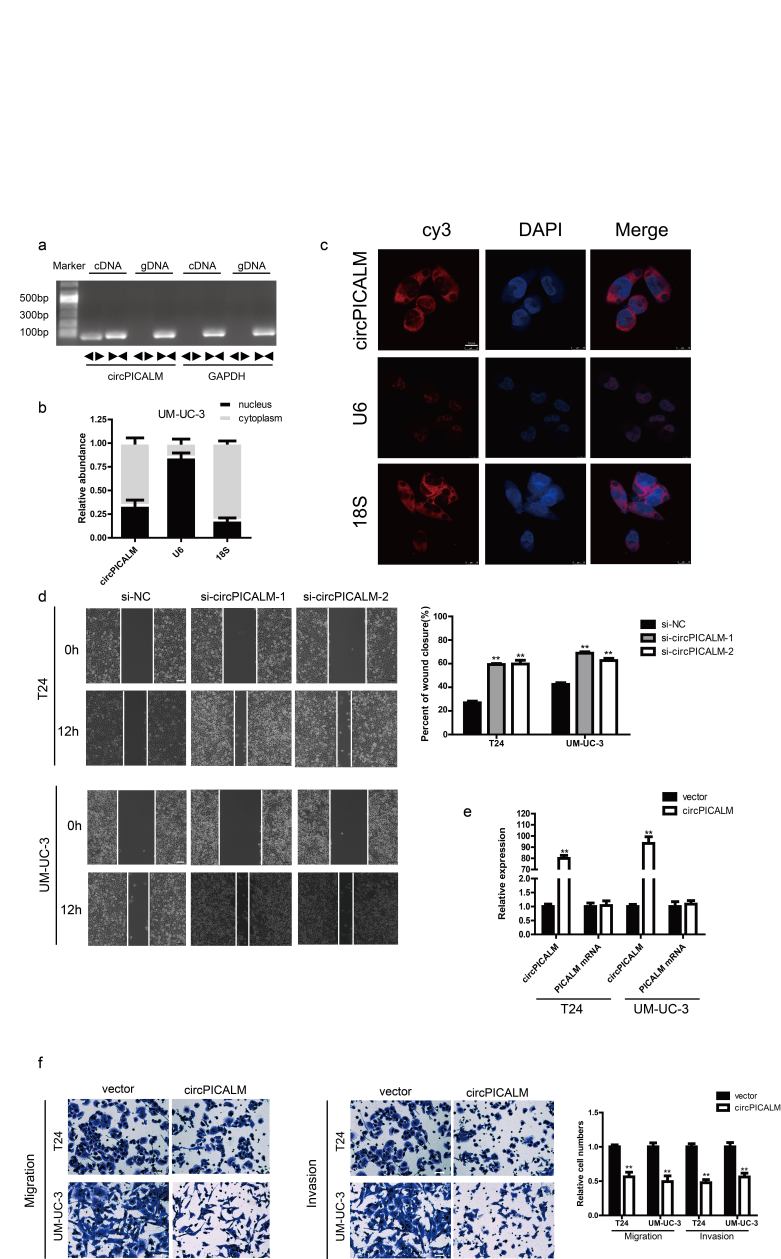
Supplementary Fig. 1.
a. PCR products of divergent or convergent primers and UM-UC-3 cDNA or gDNA were validated by gel electrophoresis. b. Subcellular distribution of circPICALM was detected by nuclear mass separation assay in UM-UC-3 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. c. Subcellular location of circPICALM was illustrated by FISH in T24 cells. U6 and 18S acted as nuclear and cytoplasm controls, respectively. Scale bar, 10 μm. d. BC cells treated with circPICALM siRNAs were measured by the wound healing assay. Scale bar, 100 μm. e. Abundance of circPICALM and PICALM mRNA in BC cells infected with NC or circPICALM overexpression lentivirus. f. Invasive abilities of BC cells overexpressing circPICALM, measured by migration and invasion assays. Scale bar, 100 μm. (Data are presented as the mean ± SD, n = 3. Unpaired, two-tailed student's t-test, **P<0.01).