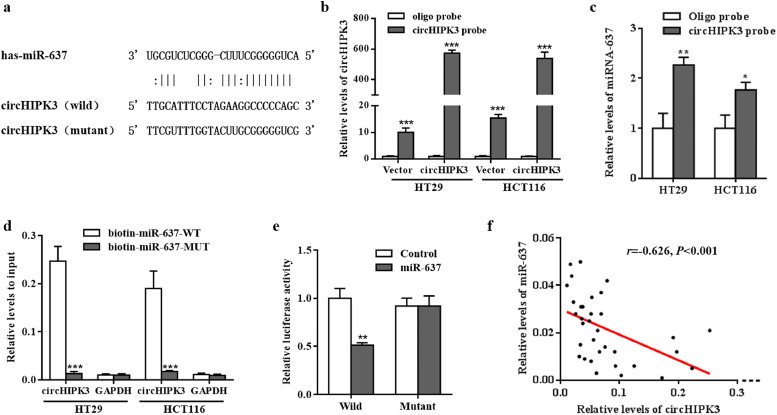
Fig. 4.
circHIPK3 functions as an efficient miR-637 sponge. (a) The putative miR-637 binding site in circHIPK3 and the corresponding mutant motif. (b) The pull-down efficiency of biotinylated-circHIPK3 probe tested by RT-qPCR in HT29 and HCT116 cells; ⁎⁎⁎P < 0•001 [student's t-test]. (c) miR-637 pull-down by biotinylated-circHIPK3 probe was tested by RT-qPCR in HT29 and HCT116 cells transfected with circHIPK3 overexpression vector; Oligo probe was used as a control; *P<0•05, ⁎⁎P < 0•01 [student's t-test]. (d) circHIPK3 pull -down by biotinylated wild-type/mutant miR-637 was tested by RT-qPCR in CRC cells with circHIPK3 overexpression. Relative levels of circHIPK3 were normalised to input; GAPDH was used as an internal control; ⁎⁎⁎P < 0•001 [student's t-test]. (e) Relative luciferase activity of wild type or mutant circHIPK3 in miR-637 mimics or controls; ⁎⁎P < 0•01 [student's t-test]. (f) Correlation between circHIPK3 and miR-637 expression in CRC tissues; r = −0•626, P < 0•001[Spearman test].