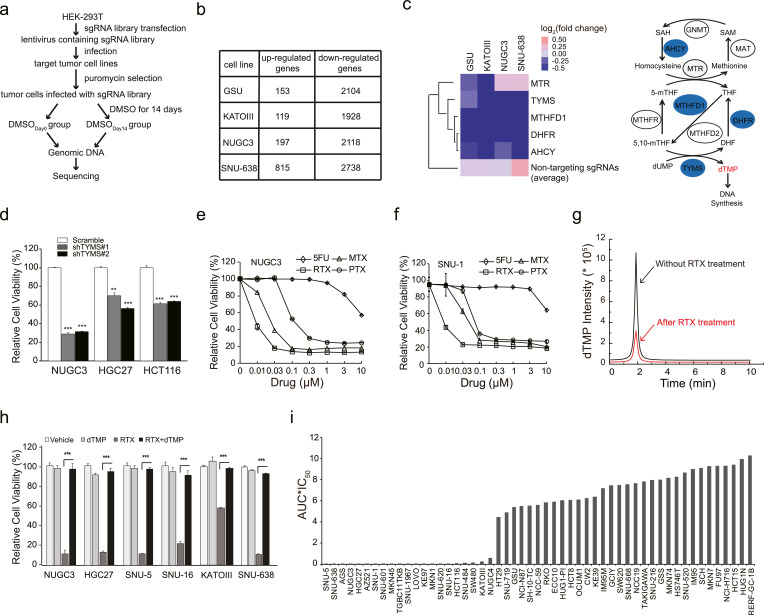
Fig. 1.
Thymidylate synthase is an important therapeutic target for treating gastrointestinal cancer. (a) The workflow of GeCKO screening in four gastrointestinal cancer cell lines. (b) The number of upregulated genes (average fold change>1.5 with duplicate sgRNAs) and downregulated genes (average fold change<0.6 with duplicate sgRNAs) in each round of GeCKO screening of each cell line. (c) Heat map of ‘one carbon pool by folate’ pathway. The average of 1,000 non-targeting sgRNA was used as control. The schematic diagram of one carbon pool by folate was shown in the right panel. Enzymes selected from the library screen data were annotated in blue circles. 5,10-methylenetetrahydrofolate reductase (MTHFR), glycine N-methyltransferase (GNMT), S-adenosylhomocysteine hydrolase (AHCY), methionine adenosyltransferase (MAT), the tri-functional C1-synthase enzyme incorporating the activities of formyl-tetrahydrofolate (THF) synthetase, cyclohydrolase and dehydrogenase activities (MTHFD1), dihydrofolate reductase (DHFR), and methylenetetrahydrofolate reductase (MTHFR). (d) The cell viability of NUGC3, HGC27 and HCT116 cells treated with RTX were detected by an MTT assay. The cell viability of NUGC3 (e) or SNU-1 (f) cells were decreased after treated with 5FU, RTX, PTX, or MTX, respectively. (g) The dTMP concentration of NUGC3 cells were detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after RTX treatment. Area under the curve (AUC) indicates the dTMP concentration. Red peak: 8 nM RTX treatment group. Black peak: 0.1% DMSO treatment group. Treatment time: 72 h. (h) dTMP efficiently rescued the cell viability of indicated cell lines when treated with RTX. Vehicle: 0.1% DMSO, RTX: 10 nM, dTMP: 20 µM. (i) The cell viability of multiple gastrointestinal cancer cell lines under the treatment of RTX. IC50 multiplied by AUC of RTX treatment was presented in this study. IC50: half maximal inhibitory concentration. AUC: area under the curve. The IC50 and AUC were calculated according to dose response curves. All the experiments were repeated at least three times, and data are represented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, p value was calculated with two-tailed Student's t-test.