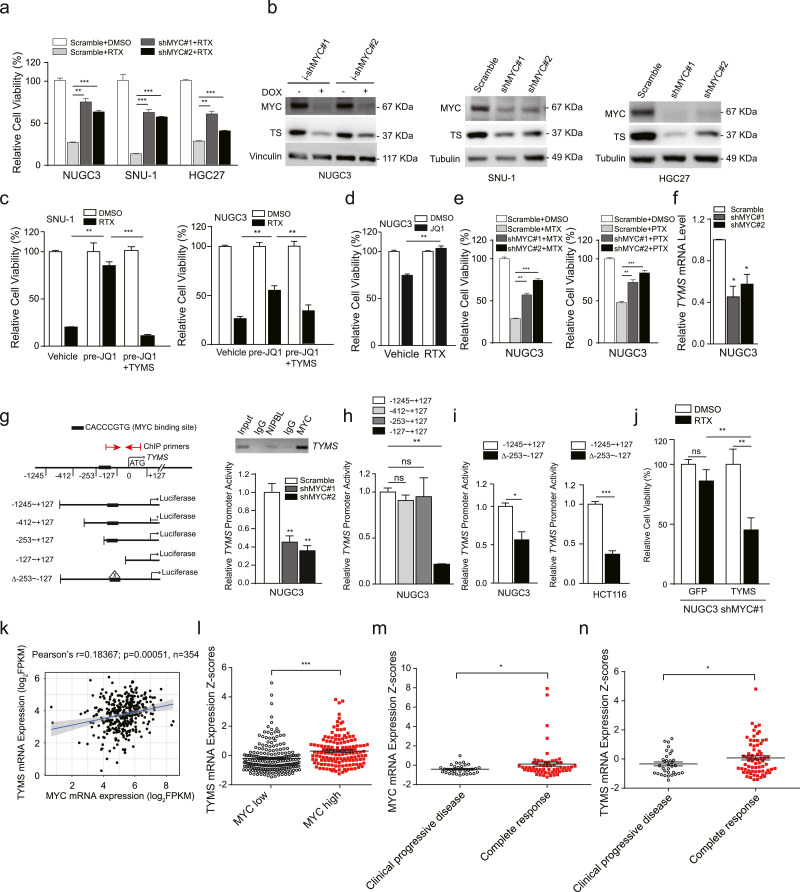
Fig. 3.
MYC predetermines the sensitivity of gastrointestinal cancer cells to antifolate drugs through regulating TYMS transcription. (a) After stably transfected with shMYC or scramble, cells were treated with RTX for three days and the cell viability was determined by an MTT assay. (b) The protein expression levels of thymidylate synthase (TS) in NUGC3, SNU-1 and HGC27 after MYC was knocked down. (c) After pre-treatment with JQ1 for 72 h, the cell viability of SNU-1 and NUGC3 cells to RTX was determined by an MTT assay, and GFP+ cell number counting was used to measure the therapeutic efficacy of RTX in TYMS-GFP transfected cells. 1 µM and 10 µM JQ1 was used for treating SNU-1 and NUGC3 cells, respectively. (d) After pre-treatment with RTX, the cell viability of NUGC3 cells to 10 µM JQ1 was determined by an MTT assay. NUGC3 cells were gradually treated with RTX from 1 nM to 90 nM in 2 months. (e) The cell viability of shMYC stably expressed NUGC3 cells was determined after treatment with MTX and PTX for three days. (f) The TYMS mRNA expression levels in NUGC3 cells after MYC knockdown, normalized with GAPDH mRNA expression levels. (g) The luciferase activity of TYMS promoter in NUGC3 cells was measured after MYC knockdown (bottom right). Results are represented as normalized relative luciferase activity with Renilla luciferase activity. MYC was immunoprecipitated with indicated DNA regions of TYMS promoter detected by ChIP-PCR (upper right). (h) The relative luciferase activity of truncated TYMS promoter fragments. (i) The relative luciferase activity of MYC binding site-deleted fragment in NUGC3 and HCT116 cells. (j) The cell viability of TYMS-GFP transfected NUGC3 shMYC cells was determined by GFP+ cell number counting after treatment with RTX. (k) Pearson correlation analysis of the correlation of the MYC mRNA expression versus the TYMS mRNA expression in the TCGA gastric cancer patient samples according to the proteinatlas website. (Pearson's r = 0.18367; p = 0.00051, n = 354). (l) The TYMS mRNA expression levels of MYC-high samples (n = 142) and MYC-low samples (n = 212), the same samples were used in (k). (m and n) The MYC (m) and TYMS (n) expression levels in gastric tumours patients who are complete response or clinical progressive disease to TYMS inhibitors in the clinical setting. The drug response data were downloaded from GDC-TCGA database, and mRNA data was retrieved from proteinatlas website. Clinical progressive disease, n = 32 (MYC), n = 33 (TYMS). Complete response, n = 59 (MYC) or n = 69 (TYMS). Data are shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, p value was calculated with two-tailed Student's t-test.