We report here the genome assembly and analysis of Microbacterium strain sp. LKL04, a Gram-positive bacterial endophyte isolated from switchgrass plants (Panicum virgatum) grown on a reclaimed coal-mining site. The 2.9-Mbp genome of this bacterium was assembled into a single contig encoding 2,806 protein coding genes.
ABSTRACT
We report here the genome assembly and analysis of Microbacterium strain sp. LKL04, a Gram-positive bacterial endophyte isolated from switchgrass plants (Panicum virgatum) grown on a reclaimed coal-mining site. The 2.9-Mbp genome of this bacterium was assembled into a single contig encoding 2,806 protein coding genes.
ANNOUNCEMENT
Members of the genus Microbacterium have previously been isolated from a wide range of environments, including soils, marine ecosystems, air, and sewage, and from plants and insects (1–5). We report here information about the sequenced and assembled genome of the bacterial endophyte Microbacterium sp. strain LKL04, a Gram-positive actinobacterium, isolated from leaves of switchgrass plants grown on a reclaimed coal-mining site in western Kentucky (6).
Switchgrass samples were collected from the coal-mining site in July 2010. Leaf samples were cut into 1- to 1.5-cm-long segments, surface sterilized with a 20% bleach solution, and rinsed 5 times with autoclaved tap water. The surface-sterilized segments were incubated on tryptic soy agar (TSA) plates for 3 to 5 days at 26°C before the individual colonies were isolated and restreaked at least three times on new TSA plates (6). Single purified colonies were then isolated and grown at room temperature for 1 to 2 days in tryptic soy broth (TSB). A modified cetyltrimethylammonium bromide (CTAB) bacterial DNA isolation protocol (7; https://1ofdmq2n8tc36m6i46scovo2e-wpengine.netdna-ssl.com/wp-content/uploads/2014/02/JGI-Bacterial-DNA-isolation-CTAB-Protocol-2012.pdf) was followed to isolate the bacterial DNA for sequencing.
The genome of Microbacterium sp. strain LKL04 was sequenced at 212× coverage using Pacific Biosciences (PacBio) sequencing technology (8). A PacBio SMRTbell library was constructed and sequenced with the PacBio RS platform, generating 198,113 filtered subreads with an average read length of 3,930 bp ± 2,621 bp, totaling 778.5 Mbp. Reads were trimmed and assembled using Hierarchical Genome Assembly Process (HGAP) v.2.3.0 (9). The final genome assembly contains a single contig spanning the complete 2.922-Mbp length of the bacterial genome, with a GC content of 69.7%, which is characteristic of actinobacteria. The genome is precited to be circular.
Genes were identified using Prodigal v.2.5, followed by a round of manual curation using GenePRIMP, resulting in a total of 2,862 predicted genes (10, 11). From these, 2,806 predicted protein coding genes were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant, UniProt, TIGRFam, Pfam, Kyoto Encyclopedia of Genes and Genomes (KEGG), Clusters of Orthologous Genes (COG), PANTHER, and InterPro databases (12–18). For the remaining 56 genes, the tRNAScan-SE tool was used to further identify 45 tRNA genes, 6 rRNA genes, and 5 noncoding RNAs. For the noncoding RNAs, the RNA components of the protein secretion complex and RNase P were identified by searching the genome for the corresponding Rfam profiles using Infernal (19, 20). CheckM v.1.0.8, hosted on KBase, was used to estimate the completeness of the LKL04 genome (21, 22). Overall, the LKL04 genome returned a completeness score of 99.5% and a contamination level of only 0.67%. Using the PANTHER hidden Markov model (HMM) scoring tool pantherScore v.2.1, the protein sequences were further mapped against the PANTHER HMM database v.14.1 to functionally annotate the LKL04 genes and query for significantly overrepresented genes (23). Default parameters were used for each software program, unless otherwise specified. Selected annotations and genome characteristics are shown in Fig. 1. Additional gene prediction analysis and manual functional annotation were performed within the Integrated Microbial Genomes (IMG) platform developed by the Joint Genome Institute (Walnut Creek, CA) (24).
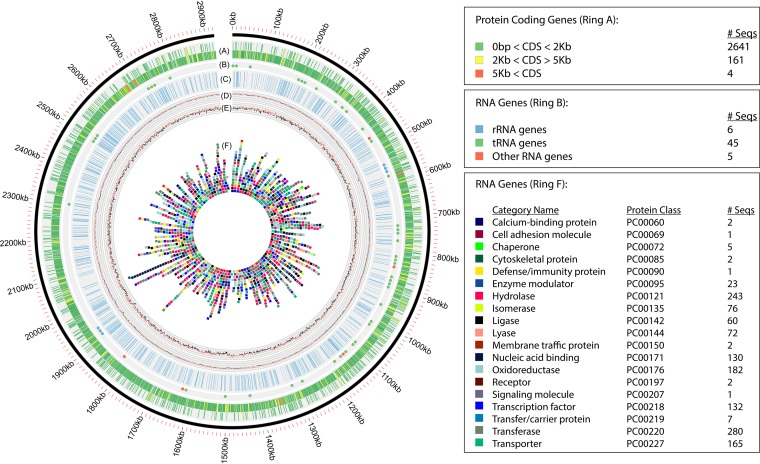
FIG 1.
Circular representation of the LKL04 genome using Circos (25). The circles, from outside to inside, denote protein coding genes colored by size (A), RNA genes (B), transmembrane helix regions (C), GC content along a 1-kb window, with red lines indicating regions above the 69.7% genome average and black lines indicating regions below the genome average (D) GC skew, with red lines indicating a skew greater than zero and black lines indicating a skew less than zero (E), and genes annotated into distinct PANTHER protein classes (F). The repository for storage of scripts used to construct the figure can be found at https://github.com/nbo245/LKL04/tree/master/circos_plot.
Data availability.
The whole-genome sequence has been deposited in DDBJ/EMBL/GenBank under the accession no. PRJNA322991. Original forward and reverse sequencing reads can be retrieved from NCBI under SRA accession no. SRR4232145 and SRR4232146. The associated sequence data can also be found at the Joint Genome Institute (JGI) portal with the IMG taxon identifier (ID) 2667527218 (https://genome.jgi.doe.gov/portal/MicspLKL04/MicspLKL04.info.html) or at https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=912630. Scripts used to construct Fig. 1 can be found at https://github.com/nbo245/LKL04/tree/master/circos_plot.
ACKNOWLEDGMENTS
Genome sequencing and data annotation were carried out in the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), a DOE Office of Science User Facility supported under contract DE-AC02-05CH11231. This work was also partially supported by the Hatch Project from USDA-NIFA-OHO01392 and by OARD Seed Award OHOA-1615 at The Ohio State University.
REFERENCES
- 1.Alves A, Correia A, Igual JM, Trujillo ME. 2014. Microbacterium endophyticum sp. nov. and Microbacterium halimionae sp. nov., endophytes isolated from the salt-marsh plant Halimione portulacoides and emended description of the genus Microbacterium. Syst Appl Microbiol 37:474–479. doi: 10.1016/j.syapm.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Kim KK, Lee KC, Oh H-M, Lee J-S. 2008. Microbacterium aquimaris sp. nov., isolated from seawater. Int J Syst Evol Microbiol 58:1616–1620. doi: 10.1099/ijs.0.65763-0. [DOI] [PubMed] [Google Scholar]
- 3.Yoon J-H, Schumann P, Kang S-J, Lee C-S, Lee S-Y, Oh T-K. 2009. Microbacterium insulae sp. nov., isolated from soil. Int J Syst Evol Microbiol 59:1738–1742. doi: 10.1099/ijs.0.007591-0. [DOI] [PubMed] [Google Scholar]
- 4.Mawlankar RR, Mual P, Sonalkar VV, Thorat MN, Verma A, Srinivasan K, Dastager SG. 2015. Microbacterium enclense sp. nov., isolated from sediment sample. Int J Syst Evol Microbiol 65:2064–2070. doi: 10.1099/ijs.0.000221. [DOI] [PubMed] [Google Scholar]
- 5.Kim DY, Shin D-H, Jung S, Kim H, Lee JS, Cho H-Y, Bae KS, Sung C-K, Rhee YH, Son K-H, Park H-Y. 2014. Novel alkali-tolerant GH10 endo-β-1,4-xylanase with broad substrate specificity from Microbacterium trichothecenolyticum HY-17, a gut bacterium of the mole cricket Gryllotalpa orientalis. J Microbiol Biotechnol 24:943–953. doi: 10.4014/jmb.1405.05032. [DOI] [PubMed] [Google Scholar]
- 6.Xia Y, Greissworth E, Mucci C, Williams MA, De Bolt S. 2013. Characterization of culturable bacterial endophytes of switchgrass (Panicum virgatum L.) and their capacity to influence plant growth. Glob Change Biol Bioenergy 5:674–682. doi: 10.1111/j.1757-1707.2012.01208.x. [DOI] [Google Scholar]
- 7.Wilson K. 2001. Preparation of genomic DNA from bacteria. Curr Protoc Mol Bio 56:241–245. doi: 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 8.Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. doi: 10.1016/j.gpb.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 10.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. 2010. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 7:455–457. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- 12.Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. 2019. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res 31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and Web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, Sneddon MW, Henderson ML, Riehl WJ, Murphy-Olson D, Chan SY, Kamimura RT, Kumari S, Drake MM, Brettin TS, Glass EM, Chivian D, Gunter D, Weston DJ, Allen BH, Baumohl J, Best AA, Bowen B, Brenner SE, Bun CC, Chandonia J-M, Chia J-M, Colasanti R, Conrad N, Davis JJ, Davison BH, DeJongh M, Devoid S, Dietrich E, Dubchak I, Edirisinghe JN, Fang G, Faria JP, Frybarger PM, Gerlach W, Gerstein M, Greiner A, Gurtowski J, Haun HL, He F, Jain R, Joachimiak MP, Keegan KP, Kondo S, Kumar V, Land ML, Meyer F, Mills M, Novichkov PS, Oh T, Olsen GJ, Olson R, Parrello B, Pasternak S, Pearson E, Poon SS, Price GA, Ramakrishnan S, Ranjan P, Ronald PC, Schatz MC, Seaver SMD, Shukla M, Sutormin RA, Syed MH, Thomason J, Tintle NL, Wang D, Xia F, Yoo H, Yoo S, Yu D. 2018. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol 36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, Thomas PD. 2019. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc 14:703. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krzywinski M, Schein H, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequence has been deposited in DDBJ/EMBL/GenBank under the accession no. PRJNA322991. Original forward and reverse sequencing reads can be retrieved from NCBI under SRA accession no. SRR4232145 and SRR4232146. The associated sequence data can also be found at the Joint Genome Institute (JGI) portal with the IMG taxon identifier (ID) 2667527218 (https://genome.jgi.doe.gov/portal/MicspLKL04/MicspLKL04.info.html) or at https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=912630. Scripts used to construct Fig. 1 can be found at https://github.com/nbo245/LKL04/tree/master/circos_plot.