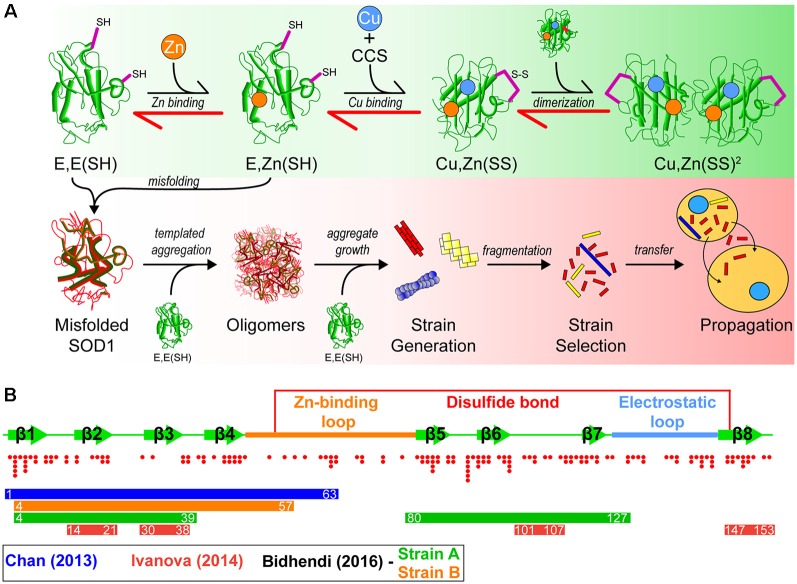
Figure 1.
A model of superoxide dismutase-1 (SOD1) aggregation and prion-like propagation. (A) The native folding (green background) and off-folding pathways (red background) of SOD1. When undergoing native folding, SOD1 is initially in a metal-free intermediate state [E, E(SH)] which is primed for Zn binding. Following Zn binding, the copper chaperone for SOD1 (CCS) heterodimerizes with a SOD1 monomer to implant Cu and facilitate the oxidation of the disulfide bond between Cys57 and Cys146 to form a mature SOD1 monomer [Cu, Zn(SS)]. Subsequent dimerization of two mature monomers gives the fully folded SOD1 enzyme. In the off-folding pathway (red background) amyotrophic lateral sclerosis (ALS)-associated mutations have the ability to push the folding back towards the nascent intermediates (red arrows), which are more aggregation prone than mature states. Misfolded SOD1 is capable of recruiting nascent SOD1 states to form oligomers. Oligomers can grow in size to form larger aggregates that may have strain-like properties. Strains that are prone to fragment are the most likely to propagate to adjacent cells and seed naïve SOD1. (B) Graphic representation of the SOD1 primary structure with important features. SOD1 is composed of 8 beta-strands and contains two major loop regions. The Zn-binding loop is responsible for coordination of Zn and, to a lesser extent, Cu. The electrostatic loop is responsible for guidance of superoxide substrate to the active site of the enzyme. A disulfide bond is formed between Cys57 and Cys146 in the mature protein. Red dots below the graphic represent the location and number of ALS-associated mutations that have been identified to date within SOD1. Coloured bars represent potential cores or contributing regions of the protein to the formation of aggregate strains from corresponding colour coded publications.