Figure 2.
Profiling Chromatin Accessibility Dynamics during Early NC Development
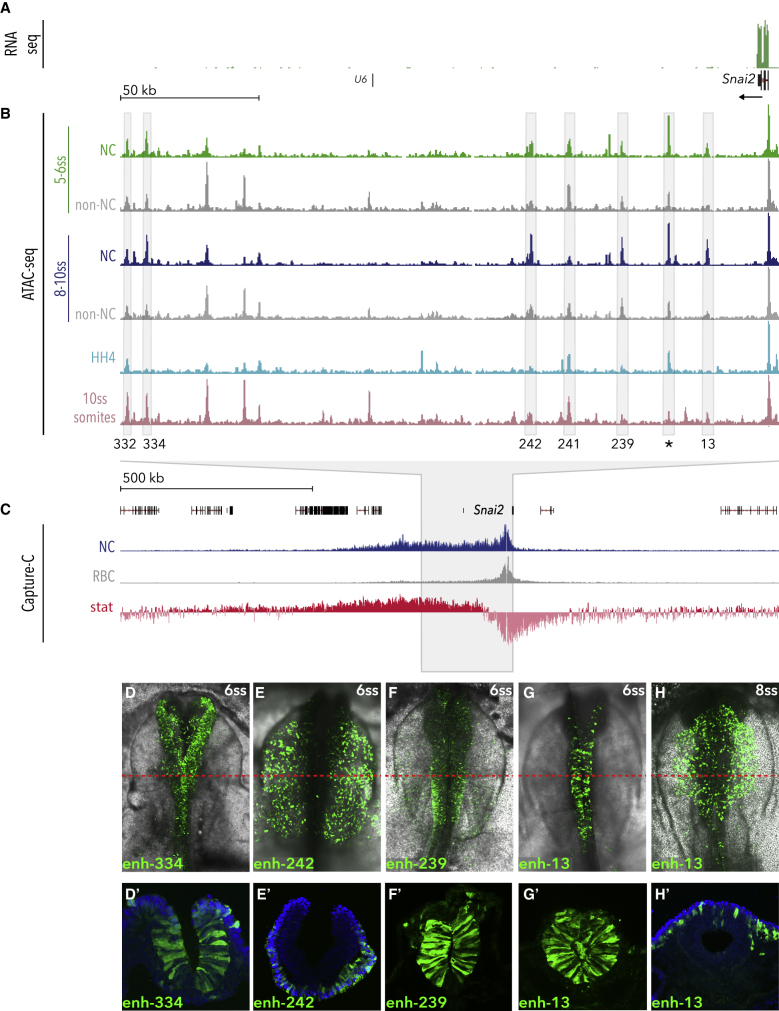
(A and B) Genome browser views of (A) 5-6ss NC RNA-seq data and (B) ATAC-seq profiles in NC and non-NC cells at the Snai2 locus. 5-6ss NC ATAC is shown in green and 8-10ss in blue; corresponding non-NC samples are shown in gray. ATAC data from HH4 and somite tissue are shown in light blue and pink, respectively (data shown is normalized to read count and shown relative to promoter peaks across all samples). Boxes indicate putative enhancers, tested in vivo.
(C) Capture-C tracks showing the TAD at the Snai2 locus in NC (blue track) and control red blood cells (RBC, gray track). Differential interactions were determined using DESeq2, hypothesis tested with Wald test and corrected for multiple testing using the Benjamin-Hochberg method. Wald statistics track (stat, in red) represents ratio of LogFoldChange values and their standard errors.
(D–H) Enhancer driven in vivo reporter (Citrine) expression of tested enhancers. (D′–H′) Transverse sections of (D)–(H), immunostained for Citrine; red line indicates approximate location of section; blue, DAPI.