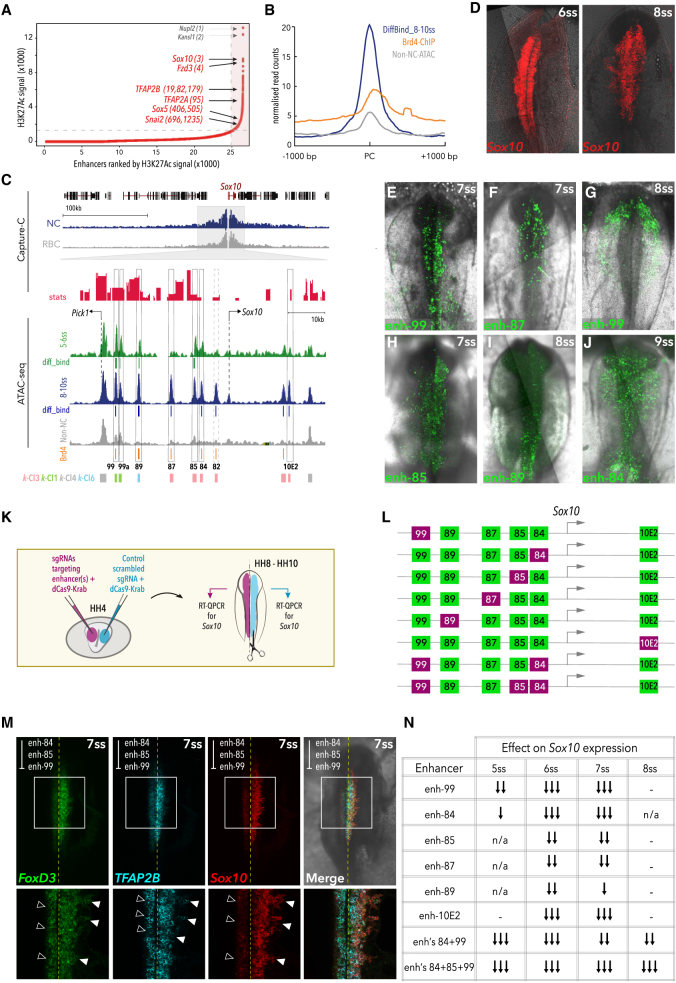
Figure 4.
Super-enhancer-like Clusters Regulate Key NC Genes
(A) Super-enhancers ranked by H3K27ac signal in 8-10ss NC, using the ROSE algorithm; top-ranked NC genes are annotated.
(B) Mean merged profile of DiffBind elements at 8-10ss and control non-NC elements occupied by Brd4.
(C) Capture-C genome browser tracks at Sox10 locus (blue, NC cells; gray, RBC; red, stat, representing ratio of LogFoldChange values and their standard errors, determined using DESeq2, p-values calculated with Wald test and Benjamin-Hochberg correction) and ATAC data (green, 5-6ss; blue, 8-10ss; gray, non-NC). DiffBind elements at 5-6ss (green) and 8-10ss (blue), Brd4-bound peaks (orange), and k-means elements are indicated. Selected putative enhancers are boxed.
(D) Endogenous Sox10 expression pattern detected by HCR.
(E–J) In vivo activity of novel Sox10 enhancers, (E) distal k-Cl1 enh-99 at 7ss, (F) enh-87 at 7ss, (G) enh-99 at 8ss, (H) enh-85 at 7ss, (I) k-Cl6 enh-89 at 8ss, (J) enh-84 at 9ss.
(K) Schematic of bilateral electroporation assay for epigenome engineering experiments.
(L) Schematic representation of decommissioned enhancers.
(M) HCR for Sox10, TFAP2B, and FoxD3 following decommissioning of targeted enhancers 84, 85, and 99.
(N) Table describing effect on endogenous Sox10 expression following epigenome modulation of Sox10 enhancers (single arrow, weak [< 20% decrease in Sox10 expression]; double arrow, moderate [20%–40% decrease]; triple arrow, strong [> 40% decrease]; n/a, not analyzed; −, no effect).