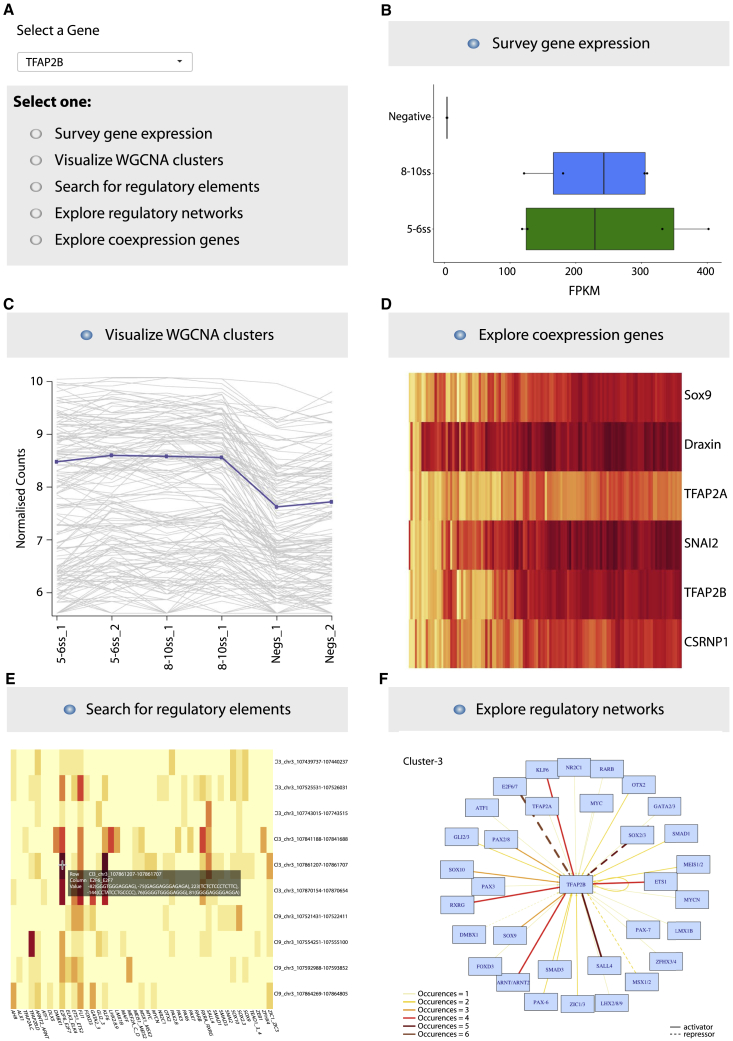
Figure 7.
Shiny App Tool for Exploration of NC Gene Expression and Regulation
(A) Searchable interface for gene of interest (GOI).
(B) Gene expression tab allows to explore levels (shown in FPKMs) as well as dynamics and differential gene expression stats across samples analyzed.
(C) WGCNA cluster tab shows assignment to co-expression clusters.
(D) Gene co-expression at the single-cell level is visualized in a searchable heatmap; features adjustable cell percentage overlap and multiple gene-name searches.
(E) Regulatory tab enables exploration of assigned CREs (genome co-ordinates in galgal4) and inherent TF motifs (exact location within element provided).
(F) Network tab enables visualization of individual regulatory circuits assembled based on either k-Cl3 and k-Cl1 enhancers and their respective TF inputs. TF occurrence frequency and activating/repressing interactions are shown.