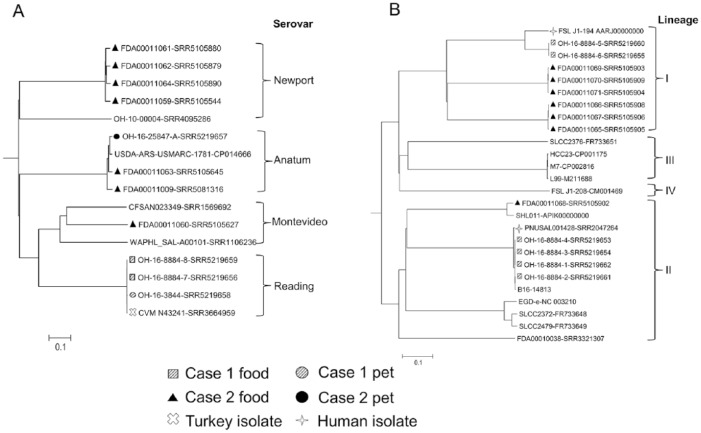
Figure 1.
Phylogenetic trees were generated for A. Salmonella enterica, B. Listeria monocytogenes, and C. Escherichia coli based on the allelic profiles of 3,288, 1,573, and 3,100 cgMLST target genes defined by SeqSphere v.3.5 (Ridom, Münster, Germany), respectively. The serovars of S. enterica and E. coli isolates were predicted based on the whole genome sequencing data using online databases.14,15,23 Scale bars indicate the distance of 10% dissimilarity.