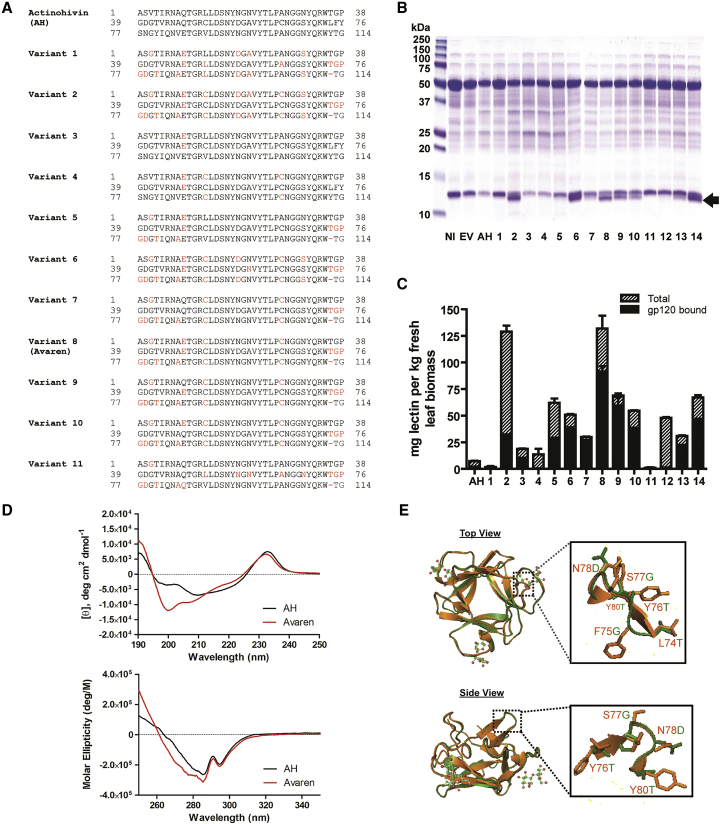
Figure 1.
Molecular Engineering of Highly Bioproducible AH Variants
(A) The amino acid sequences of AH (UniProt: Q9KWN0) and AH variants. The three domains of mature AH (amino acids 1–38, 39–76, and 77–114) are aligned. Variant 8 was designated as Avaren (see Results). (B) SDS-PAGE was performed to analyze crude extracts of N. benthamiana leaves expressing AH or its variants and stained with Coomassie brilliant blue. At 5 dpi, leaf proteins were extracted with PBS (pH 7.2) containing 40 mM ascorbic acid using a 3:1 buffer-to-leaf ratio. NI, non-infiltrated leaf extract; EV, empty-vector-infiltrated leaf extract; AH, AH-expressing leaf extract. Lanes 1–14: leaf extracts of AH variants. (C) Quantification of AH variants in N. benthamiana leaf tissue using direct and gp120-capture ELISA for total protein (hatched bars) and gp120-binding protein (black bars) detected by a rabbit anti-AH antiserum. Variant 8 was designated as Avaren. (D) CD analysis. Far-UV CD (top) and near-UV CD (bottom) spectra of AH and Avaren. (E) Crystal structure of AH (PDB: 4G1R) (orange) superimposed with a homology model of Avaren (green), shown from the top and side views (PyMOL software). Homology modeling was performed with SWISS-MODEL, using AH as a template. Zoomed images of the surface-exposed loop between domains 2 and 3 (amino acids 74–80) are boxed.