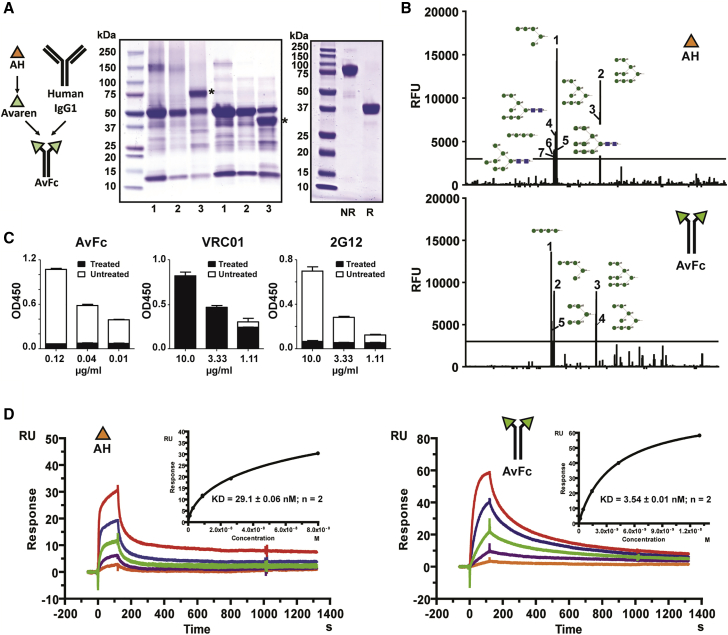
Figure 2.
Design, Production, and HMG-Binding Profiles of AvFc
(A) Expression and purification of AvFc. Reducing SDS-PAGE and non-reducing SDS-PAGE were performed to analyze crude leaf extracts and purified AvFc, stained with Coomassie brilliant blue. Representative gel images are shown. Left: 1, non-infiltrated leaf extract; 2, empty-vector-infiltrated leaf extract; 3, AvFc-expressing leaf extract. Asterisks indicate AvFc. Right: purified AvFc under non-reducing and reducing conditions. NR, non-reducing conditions. R, reducing conditions. (B) Glycan array analysis. Sugar-binding profiles of AH (top) and AvFc (bottom) were analyzed in a mammalian glycan array with 610 glycans by the Consortium for Functional Glycomics. Glycans with a mean relative fluorescence unit exceeding 3,000 were ranked and indicated with schematic diagrams. Green circles indicate mannose; blue squares indicate N-acetylglucosamine. See Data Availability for a complete dataset. (C) Analysis of AvFc’s binding to gp120 treated with α-mannosidase. The gp120-binding ELISA was performed on AvFc, VRC01 (a HIV-1 CD4 binding-site-specific monoclonal antibody), and 2G12 (an HIV-1 envelope HMG-binding monoclonal antibody), using a recombinant HIV-1 gp120 or gp120 treated with α (1-2,3,6) mannosidase. The gp120-binding activities of AvFc and 2G12, but not of VRC01, were abolished by treating the Env protein with α-mannosidase, demonstrating AvFc’s specificity to the terminal mannose residues of HMGs. (D) SPR analysis of the binding affinities of AH and AvFc to gp120SF162. The binding kinetics and affinities of AH (left) and AvFc (right) to gp120SF162 were measured using a Biacore X100 2.0 instrument at ambient temperature. Representative sensorgrams are shown. A recombinant gp120SF162 was captured on a sensor chip to a surface density of about 50–100 RU. 3-fold serial dilutions of AvFc (1 μg/mL to 0.0123 μg/mL) or AH (1 μg/mL to 0.0123 μg/mL) were injected at a flow rate of 5 μL/min. KD was determined based on steady state (inset). Data indicate mean ± SEM from two independent analyses.