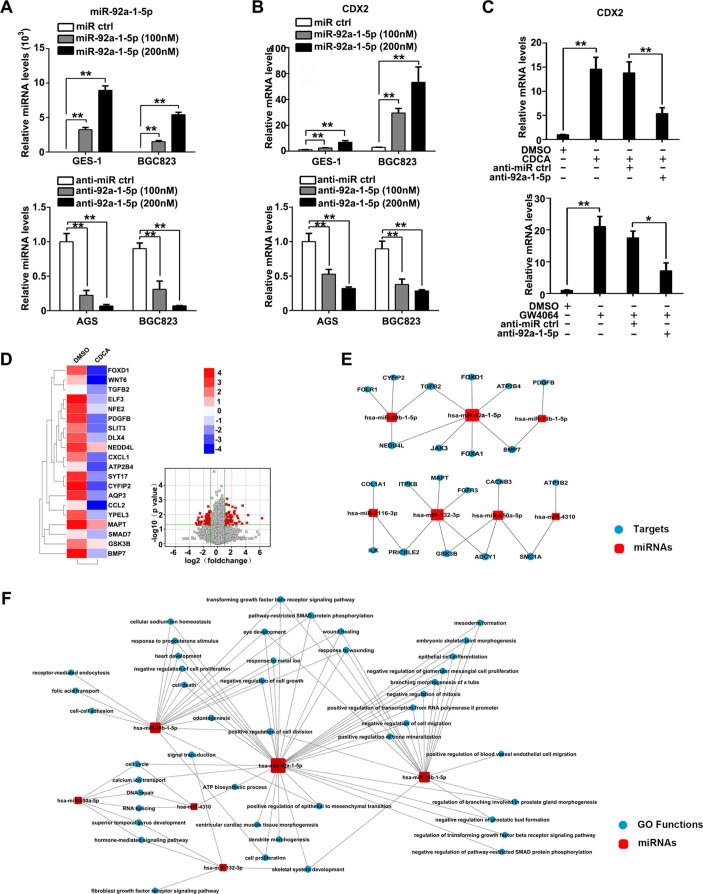
Figure 3.
miR-92a-1–5p increases expression of CDX2 and intestinal markers and miR-92a-mRNA regulatory networks. (A) Gastric epithelial cell line (GES)-1 and BGC823 cells were transfected with miR-92a-1–5p mimics or inhibitors at a final concentration of 100 and 200 nM. miR-92a-1–5p expression was detected by qRT-PCR at 48 hours post-transfection. U6 was used as an internal control in qRT-PCR of miR-92a-1–5p. (B) GES-1, BGC823 and AGS cells were transfected as in figure 3A and CDX2 expression was examined by qRT-PCR. (C) GES-1 cells was treated with CDCA (top) or GW4064 (bottom) together with inhibitor of miR-92a-1–5p and CDX2 expression was determined by qRT-PCR. GAPDH RNA was used as internal control in qRT-PCR of CDX2. (D) Whole genome expression profiles for GES-1 cells treated with CDCA (100 µM) for 24 hours. Heat map (left) and Volcano plot (right) illustrating the global differences in gene expression between CDCA-treated GES-1 cells and control (fold change >2.0; p<0.05). (E) Bioinformatic analysis of microRNA (miRNA)-targets regulation through integrating miRNA microarray results and whole genome expression profiles results. Red square: increased miRNAs in CDCA-treated GES-1 cells. Blue circles: decreased genes in CDCA-treated GES-1 cells. (F) GO analysis of increased miRNAs in CDCA-treated GES-1 cells focusing on miR-92a-1–5p function. Means±SEM of a representative experiment (n=3) performed in triplicates are shown. *P<0.05; **p<0.01.