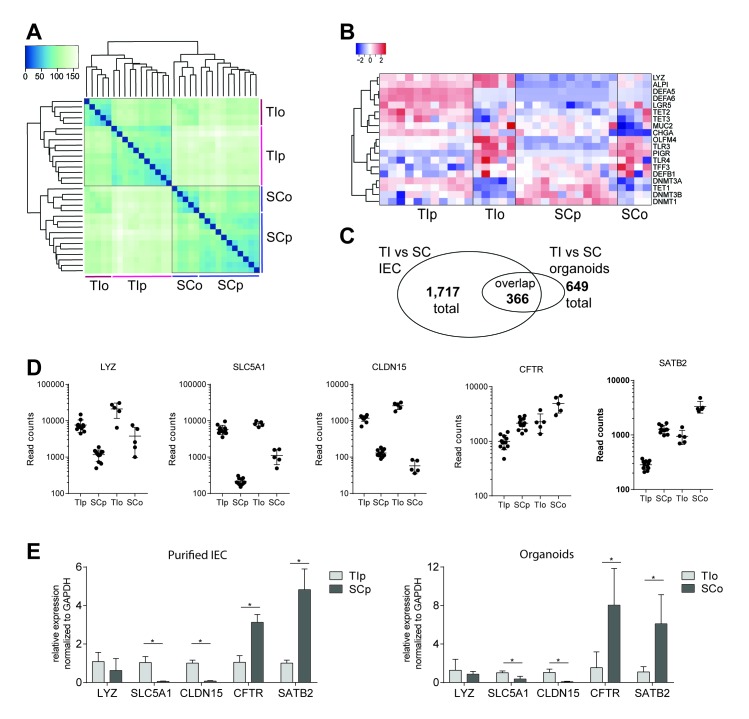
Figure 2.
Transcriptomic profiling of human intestinal epithelial organoids (IEOs) and primary intestinal epithelial cells (IECs). (A) Hierarchical clustering and sample heatmap of transcriptomes by RNA-sequencing of IECs and organoids from terminal ileum (TI) and sigmoid colon (SC). (B) Heatmap displaying gene expression (ie, rlog-transformed RNA-seq counts) of selected epithelial cell subset markers, genes involved in intestinal epithelial innate immune function and regulation of DNA methylation. (C) Venn diagram displaying number as well as overlap of differentially expressed genes (DEGs) comparing TI versus SC of primary IEC and organoids. Significance cut-off adj. p<0.01 and log2Fold Change>±1.5, n=11 (IEC) and 5 (IEO) for each gut segment. Organoids were passages 1–6. (D) RNA-seq read counts of selected marker genes found to retain gut segment-specific expression patterns in organoid culture. (E) Validation of differentially expressed marker genes by quantitative PCR on a validation sample set, represented as mean+SD, n=3–5 per group, *p<0.05. ALPI, intestinal alkaline phosphatase; CFTR, cystic fibrosis-transmembrane conductance regulator; CHGA, chromogranin A; CLDN15, claudin 15; DEFA, defensin alpha; DEFB1, defensin beta 1; DNMT, DNA methyltransferase; LGR5, leucin-rich repeat containing G protein-coupled receptor 5; LYZ, lysozyme; MUC2, mucin 2; OLFM4, olfactomedin-4; PIGR, polymeric immunoglobulin receptor; SATB2, special AT-rich sequence-binding protein 2; SLC5A1, solute carrier family 5 member 1; TET, ten-eleven translocation; TFF3, trefoil factor 3; TLR, Toll- like receptor.