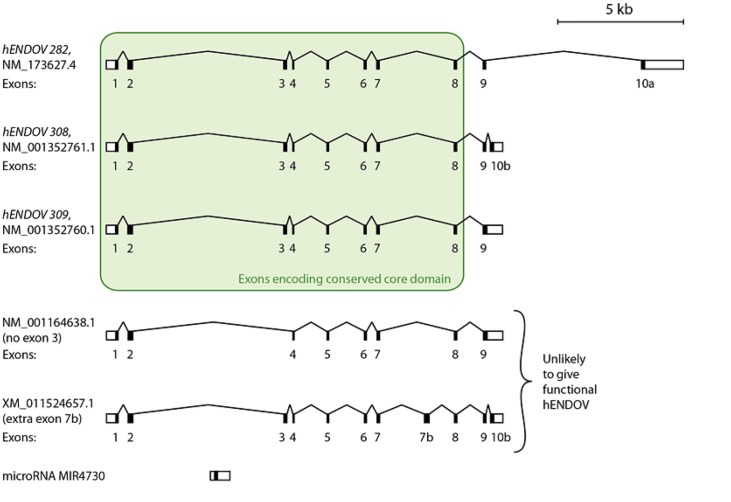
Fig 1. Transcript variants of hENDOV.
Among more than 40 predicted transcripts for hENDOV, only three; hENDOV 282 (NCBI RefSeq identifier NM_173627.4), hENDOV 308 (NM_001352761.1) and hENDOV 309 (NM_001352760.1) comprise all exons encoding the conserved catalytic core (green box, exons 1 to 8) and no more than one or two additional 3’ exons. The two lower transcripts shown in the figure are examples of other transcripts that either lack important exons encoding the catalytic core, or have additional non-conserved exons encoding segments that are likely to disrupt the structure and function of the protein. The hENDOV gene is located on chromosome 17q25.3, on the positive strand, with a length of approximately 25,000 base pairs (see scale bar above the gene structure). The lengths of the exons are not shown at the correct scale. 5’-untranslated regions (5’-UTRs) and 3’-UTRs are shown as white boxes in exon 1 and the most 3’ exon, respectively, for each transcript. MIR4730, a non-coding RNA in the microRNA class, transcribed in the same direction as hENDOV, is located in intron 2 (lowest transcript).