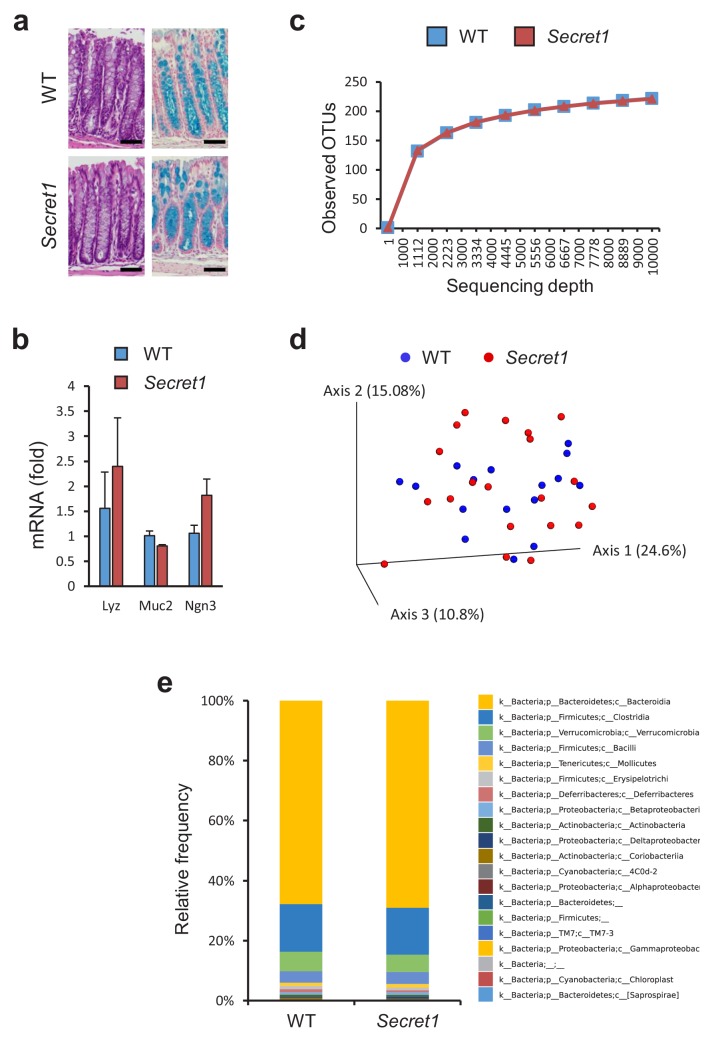
Figure 4. Scgn deficient mice display intact mucosal architecture and microbiota at baseline.
(a) Representative colon histologic images from Secret1 and WT animals under untreated conditions. HE is shown on the left, alcian blue staining on the right. Scale bar 100 µm. (b) Baseline expression of prototypical intestinal epithelial cell lineage-specific markers from small bowel in Secret1 and WT mice was determined by qRT-PCR (n = 3 in each group). (c) Microbiome alpha diversity as measured by observed OTU mean counts of fecal 16 s rRNA sequencing. (d) Beta diversity by unweighted UNIFRAC principal coordinate analysis (PCoA) of fecal 16 s rRNA sequencing. (e) Class level taxonomic composition for stool 16 s sequencing. WT n = 16 Secret1 n = 20. S.E.M. was used for error bars in (b).