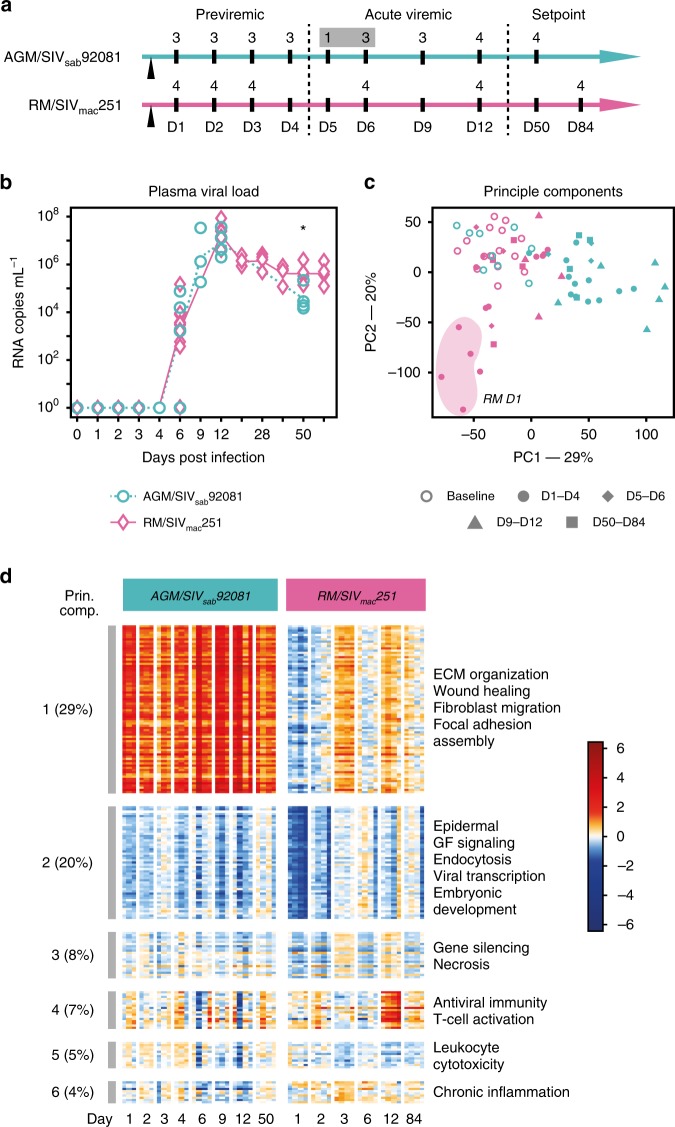
Fig. 1.
Study overview. a Study design. Numbers indicate the number of necropsied animals at each time point. b Plasma viral load in AGMs and RMs, given as log10 values. Significant difference (p < 0.05, two-sided T-test) is indicated by a star. c Principle component (PC) analysis of the 7420 DE genes, showing the strongest contrasts between AGM and RM time dynamics. Numbers at the axis indicate the percentage of total variation explained. d Heatmap showing representative gene sets obtained by PC analysis. From the top six principal components, we show the genes with the strongest contribution to variation in proportional numbers. A functional enrichment test was performed on these gene sets; the top enriched functions are listed on the right of the heatmap