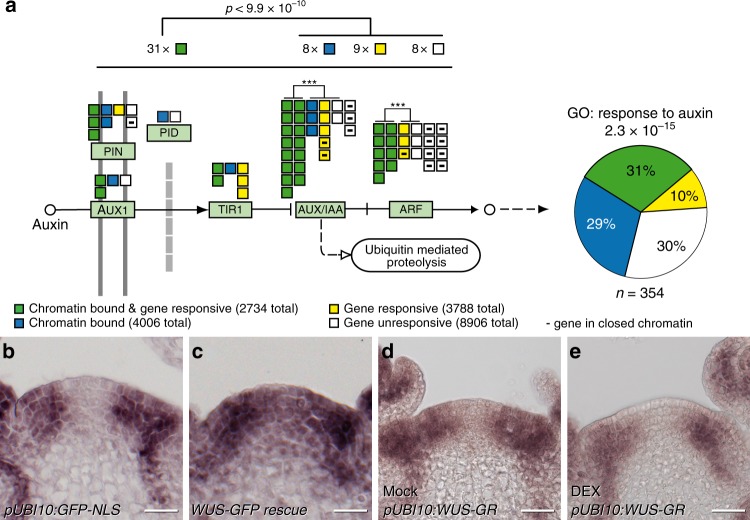
Fig. 5.
Pathway level control underlies WUSCHEL mediated gating of auxin signaling. a RNA-seq and ChIP-seq data demonstrate that WUS globally affects the auxin pathway, including transport, perception, signal transduction, as well as transcriptional response. Genes involved in the auxin signaling pathway are depicted as color-coded boxes, data on WUS binding and transcriptional response are shown in Supplementary Table 2. Across the entire pathway, WUS bound and responsive genes are overrepresented (p < 9.9 × 10−10 Fisher exact test for 2 × 2 matrix, bound and responsive genes (green color code) vs. all other classes, open chromatin as a background). Even within gene families, such as AUX/IAA or ARF, WUS targets are overrepresented (***p < 10−4 by Fisher exact test for 2 × 2 matrix, bound and responsive genes (green color code) vs. all other classes, open chromatin as a background). Genes downstream this pathway are annotated as “responsive to auxin” and are also highly overrepresented among WUS targets (GO term enrichment taken from Supplementary Table 1). b, c MP RNA accumulation 24 h post anti-GFP nanobody induction in a pUBQ10:GFP-NLS control line (b) and the pWUS:WUS-linker-GFP wus rescue background (c). d, e Response of MP mRNA to ectopic activation of WUS-GR. MP RNA after 24 h of mock (d) or DEX treatment (e). Scale bars 20 µm