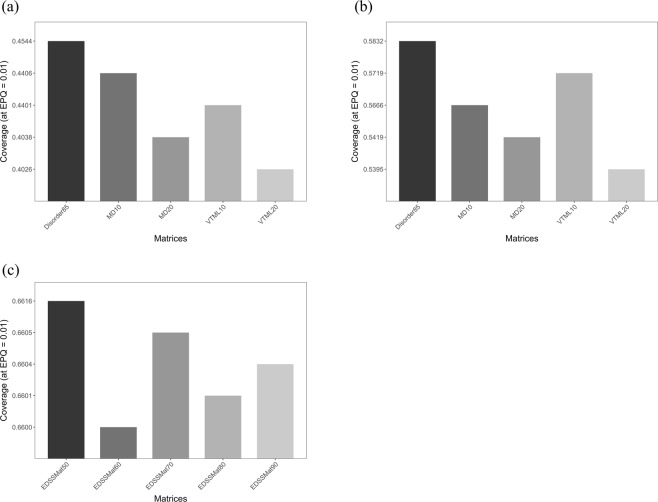
Figure 2.
Relative entropy-independent comparison of top 5 search matrices for homology detection using three test datasets: (a) Less Disordered (LD); (b) Moderately Disordered (MD); and (c) Highly Disordered (HD). Quadratically normalised coverage measure (Qquad) at 0.01 errors per query (EPQ) on y axis reports the fraction of true positive family relations at a restricted number of false positives. Height of a bar in the figure represents coverage (Qquad) achieved by a matrix. All EDSSMat series of matrices achieved higher coverage values (Qquad) than other comparing matrices on HD test dataset. On MD and LD test datasets, along with Disorder85, lower numbered MD and VTML search matrices are the best performers. Differences in coverage measure are also statistically significant as |Z| ≥ 1.96 (Supplementary Tables 6–8).