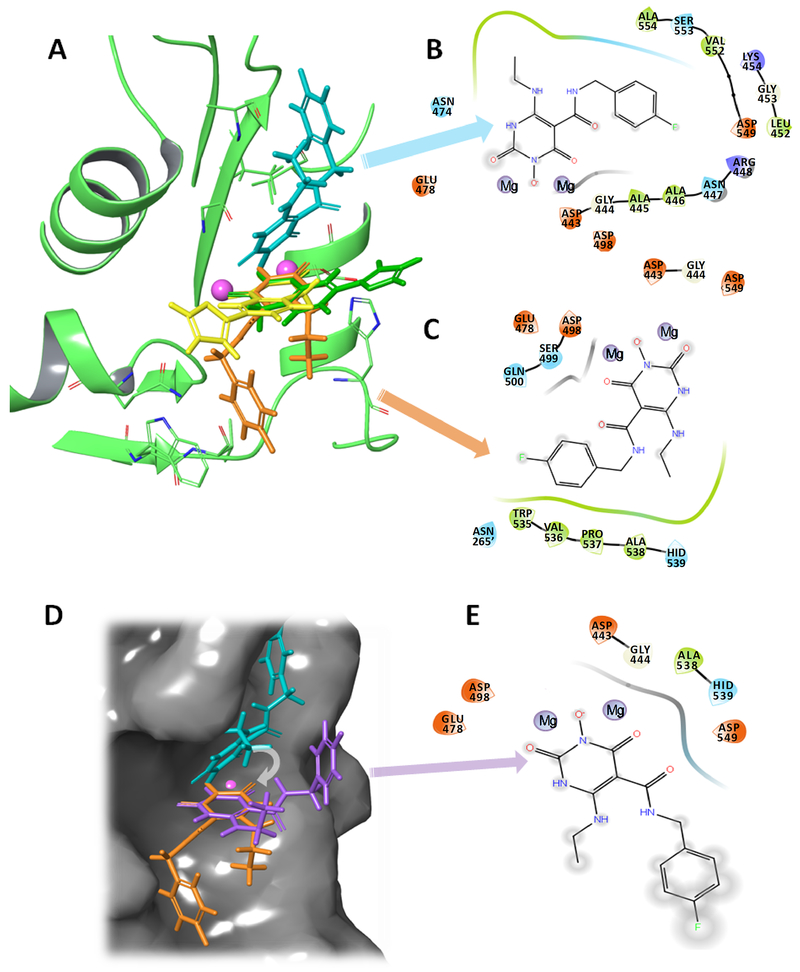
Figure 8.
The two binding modes of ZW566 are distinctive from the two-mode binding modes of YLC2-155. A) Overlap of ZW566 (cyan and orange structures) and YLC2-155 (yellow and green structures) binding modes. B) Ligand interaction diagram (LID) for ZW566 binding mode 1. C) LID for ZW566 binding mode 2. D) ZW566 binding mode 1 (cyan structure), mode 2 (orange structure) and a simulated “YLC-2155-like binding mode” for ZW566 (purple structure) obtain by superposition of the ZW566 chelating ring in binding mode 1 with the ZW566 chelating ring in binding Mode 2. Structures are shown on the RNH active site surface. E) LID for ZW566 superimposed to simulate YLC2-155 binding mode. Such a binding mode would result in fewer interactions with amino acid residues and greater exposure to solvent. In LID, negatively charged residues are shown in red, positively charged residues are shown in blue, hydrophobic residues are shown in green, polar residues are shown in cyan, and glycine is shown in white. Note, in the LID presentation, residue numbers were embedded into the original graphics. In panel C, dashed residue number indicates that from p51 subunit in RT.