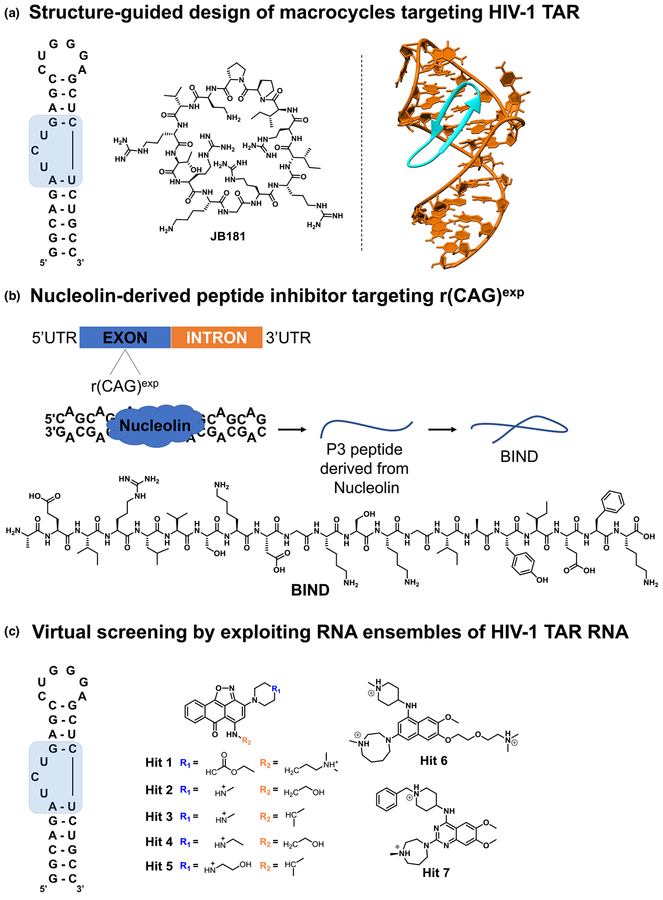
FIGURE 1.
Graphical representations of the structure-based design strategies used to identify small molecules interacting with RNA. (a) Macrocycle JB181 was generated from L22 peptide via a focused positional scanning strategy with noncanonical amino acids. The RNA motif targeted by JB181 is highlighted in blue. (b) The binding interface of the Nucleolin–r(CAG)exp complex was used to generate peptide P3, which was subsequently optimized to adopt a b-hairpin conformation as in Beta-structured Inhibitor for Neurodegenerative Diseases (BIND). (c) Chemical structures of small-molecule hit compounds identified by a Tat-peptide displacement assay and confirmed by virtual screening using an ensemble-based virtual screening strategy.