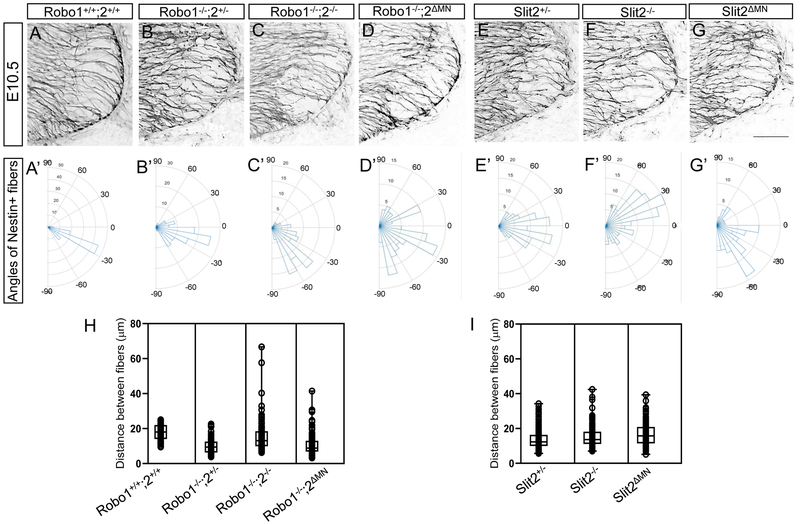
Figure 5. Nestin-positive neuroepithelial cells are disorganized in Robo or Slit2 knockout spinal cords.
(A-G) Nestin labeling on E10.5 (n=6 embryos for each genotype) spinal cord sections in Robo (A-D) and Slit (E-G) embryos showing that Nestin+ neuroepithelial/radial glial processes were sparse, disorganized, and disrupted in the ventral spinal cord of Robol−/−;2−/−, Robo1−/−;2ΔMN, Slit2−/− or Slit2ΔMN embryos. embryos. (A’-G’) Rose histograms showing the distributions of angles inside the spinal motor column. Angles of each projection were clustered in 10° bins. The length of the radius of each segment represents the percentage of total fibers include each bin. Note that the radial lengths were shown on a shorter scale in the mutant graphs because the angles were more variable than wild-type embryos, and therefore had lower percentages per bin. (H, I) Nested box-and-whisker plots with aligned data points showed that distance between Nestin+ processes has a high variance in Robo1−/−;2−/− or Robo1−/−;2ΔMN, Slit2−/− or Slit2ΔMN embryos compared to wild-type or their littermate controls. Scale bars: A-F, 50 μm.*: P <0.05, **: P <0.001.