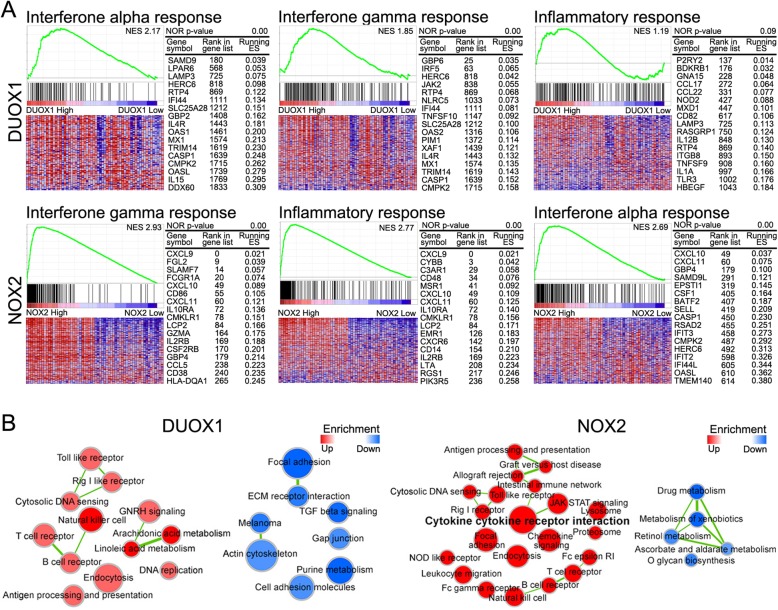
Fig. 3.
Gene set enrichment analysis and map visualization for DUOX1 and NOX2 in cervical cancer. a Representative GSEA data with p values for DUOX1 and NOX2 was shown. b Enrichment maps of DUOX1 and NOX2 in KEGG pathways. Red nodes represent enrichment in the former, whereas blue nodes represent enrichment in the latter. Color intensity is proportional to the degree of enrichment, and clusters represent functionally related gene sets. Data are for the 10% of samples with the most strongly upregulated DUOX1 and NOX expression and the 10% of samples with the most strongly downregulated DUOX1 and NOX expression. The NES (Normalized Enrichment Score) computes the density of modified genes in the dataset with the random expectancies, normalized by the number of genes found in a given gene cluster, to take into account the size of the cluster