Fig. 3.
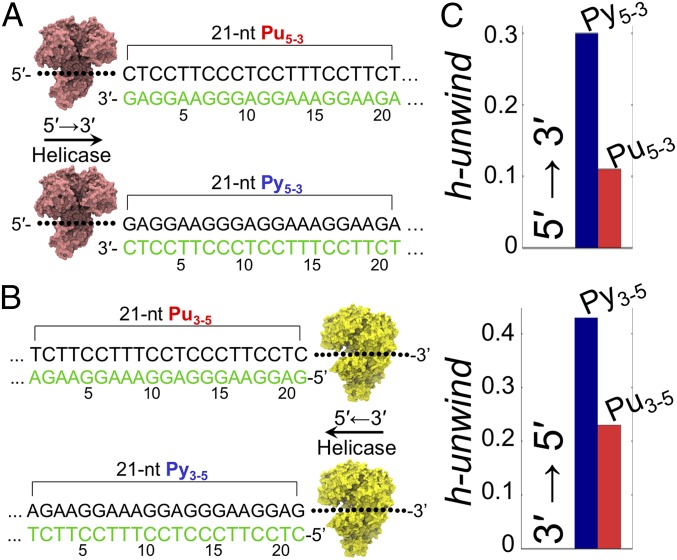
The h-unwind predicts a direction-dependent unwinding efficiency of helicases. (A) DNA duplexes Pu5-3 and Py5-3 have a 21-nt sequence in the displaced strand (in green) that is homopurine and homopyrimidine, respectively. (B) Pu3-5 and Py3-5 have a homopurine and homopyrimidine sequence in the displaced strand (in green), respectively. (C) Duplexes with homopyrimidines in the displaced strand (Py5-3 and Py3-5) have higher h-unwind values and are thus predicted to be more efficiently unwound than the homopurine analogs (Pu5-3 and Pu5-3).