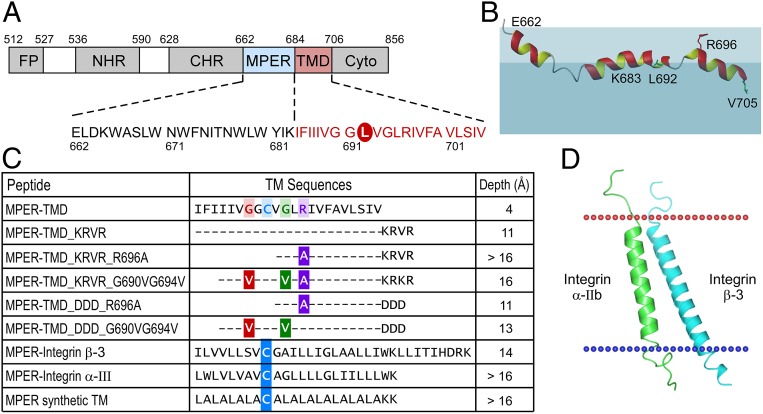
Fig. 1.
Effect of cytoplasmic and TM sequences on the immersion depth of MPER-TMD in a membrane environment. (A) Domain architecture of gp41. Cyto, cytoplasmic domain; FP, fusion peptide. The peptide sequence used in this study comprised a L692C mutation for R1 spin labeling circled in red. (B) NMR structure of the HxB2 MPER-TMD segment simulated in the liposome (43). The colored region representing the lipid membrane is for illustration only. Lipid headgroup is represented in light blue and aliphatic region is shaded in darker blue. R1 spin labels are placed at the position of green L692 and V705. R696 side-chain position is shown in red relative to the lipid headgroup. (C) Immersion depths of L692 residue in various MPER-TMD segments from EPR measurements. Mutations are color-coded on the wild-type sequence. Cysteine mutations for R1 spin labeling in the MPER-TMD chimeras are highlighted in blue. (D) A model structure of TMD from integrin α-IIb (green) and from integrin β-3 (cyan) used for chimeric MPER-TMD peptide. The models were adapted from 2K9J of the Orientations of Proteins in Membranes (OPM) database.